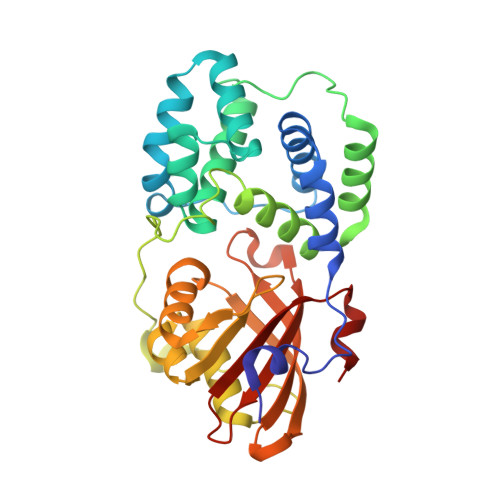
The crystal structure of a cyanobacterial water-soluble carotenoid binding protein.
Kerfeld, C.A., Sawaya, M.R., Brahmandam, V., Cascio, D., Ho, K.K., Trevithick-Sutton, C.C., Krogmann, D.W., Yeates, T.O.(2003) Structure 11: 55-65
- PubMed: 12517340
- DOI: https://doi.org/10.1016/s0969-2126(02)00936-x
- Primary Citation of Related Structures:
5UI2 - PubMed Abstract:
Carotenoids undergo a wide range of photochemical reactions in animal, plant, and microbial systems. In photosynthetic organisms, in addition to light harvesting, they perform an essential role in protecting against light-induced damage by quenching singlet oxygen, superoxide anion radicals, or triplet-state chlorophyll. We have determined the crystal structure of a water-soluble orange carotenoid protein (OCP) isolated from the cyanobacterium Arthrospira maxima at a resolution of 2.1 A. OCP forms a homodimer with one carotenoid molecule per monomer. The carotenoid binding site is lined by a striking number of methionine residues. The structure reveals several possible ways in which the protein environment influences the spectral properties of the pigment and provides insight into how the OCP carries out its putative functions in photoprotection.
- Molecular Biology Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA. kerfeld@mbi.ucla.edu
Organizational Affiliation: