The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234
Tan, K., Li, H., Endres, M., Phillips Jr., G.N., Joachimiak, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glyoxalase/bleomycin resisance protein/dioxygenase | 129 | Streptomyces sp. CB03234 | Mutation(s): 0 Gene Names: tnmS1 | 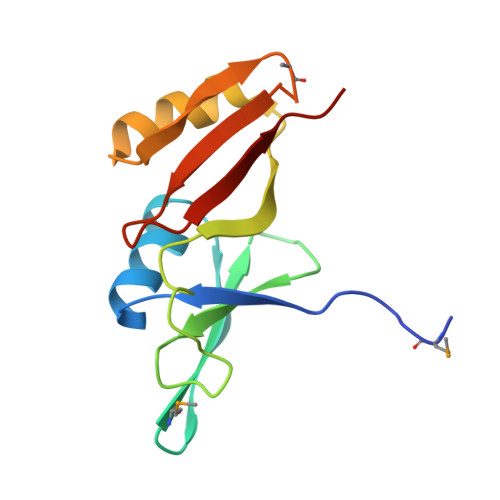 | |
UniProt | |||||
Find proteins for A0A125SA24 (Streptomyces sp. (strain CB03234)) Explore A0A125SA24 Go to UniProtKB: A0A125SA24 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A125SA24 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FMT Query on FMT | C [auth A] | FORMIC ACID C H2 O2 BDAGIHXWWSANSR-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 82.836 | α = 90 |
| b = 33.346 | β = 98.49 |
| c = 101.781 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| HKL-3000 | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM115586 |