The Loss of Expression of a Single Type 3 Effector (CT622) Strongly ReducesChlamydia trachomatisInfectivity and Growth.
Cosse, M.M., Barta, M.L., Fisher, D.J., Oesterlin, L.K., Niragire, B., Perrinet, S., Millot, G.A., Hefty, P.S., Subtil, A.(2018) Front Cell Infect Microbiol 8: 145-145
- PubMed: 29868501
- DOI: https://doi.org/10.3389/fcimb.2018.00145
- Primary Citation of Related Structures:
5UE0 - PubMed Abstract:
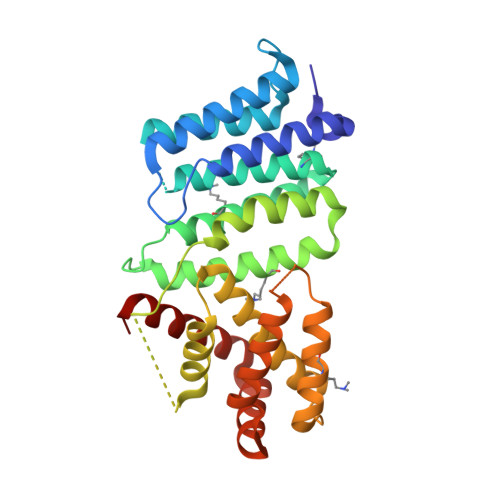
Invasion of epithelial cells by the obligate intracellular bacterium Chlamydia trachomatis results in its enclosure inside a membrane-bound compartment termed an inclusion. The bacterium quickly begins manipulating interactions between host intracellular trafficking and the inclusion interface, diverging from the endocytic pathway and escaping lysosomal fusion. We have identified a previously uncharacterized protein, CT622, unique to the Chlamydiaceae , in the absence of which most bacteria failed to establish a successful infection. CT622 is abundant in the infectious form of the bacteria, in which it associates with CT635, a putative novel chaperone protein. We show that CT622 is translocated into the host cytoplasm via type three secretion throughout the developmental cycle of the bacteria. Two separate domains of roughly equal size have been identified within CT622 and a 1.9 Å crystal structure of the C-terminal domain has been determined. Genetic disruption of ct622 expression resulted in a strong bacterial growth defect, which was due to deficiencies in proliferation and in the generation of infectious bacteria. Our results converge to identify CT622 as a secreted protein that plays multiple and crucial roles in the initiation and support of the C. trachomatis growth cycle. They reveal that genetic disruption of a single effector can deeply affect bacterial fitness.
- Unité de Biologie Cellulaire de l'Infection Microbienne, Institut Pasteur, Paris, France.
Organizational Affiliation: