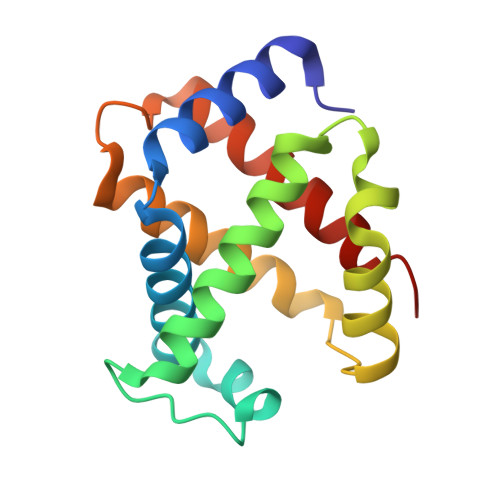
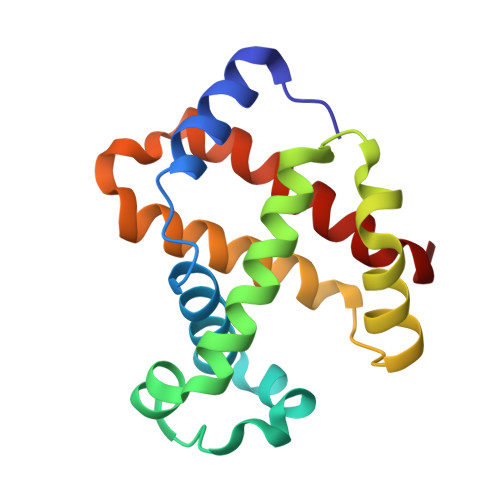
Structural and Mechanistic Insights into Hemoglobin-catalyzed Hydrogen Sulfide Oxidation and the Fate of Polysulfide Products.
Vitvitsky, V., Yadav, P.K., An, S., Seravalli, J., Cho, U.S., Banerjee, R.(2017) J Biological Chem 292: 5584-5592
- PubMed: 28213526
- DOI: https://doi.org/10.1074/jbc.M117.774943
- Primary Citation of Related Structures:
5UCU - PubMed Abstract:
Hydrogen sulfide is a cardioprotective signaling molecule but is toxic at elevated concentrations. Red blood cells can synthesize H 2 S but, lacking organelles, cannot dispose of H 2 S via the mitochondrial sulfide oxidation pathway. We have recently shown that at high sulfide concentrations, ferric hemoglobin oxidizes H 2 S to a mixture of thiosulfate and iron-bound polysulfides in which the latter species predominates. Here, we report the crystal structure of human hemoglobin containing low spin ferric sulfide, the first intermediate in heme-catalyzed sulfide oxidation. The structure provides molecular insights into why sulfide is susceptible to oxidation in human hemoglobin but is stabilized against it in HbI, a specialized sulfide-carrying hemoglobin from a mollusk adapted to life in a sulfide-rich environment. We have also captured a second sulfide bound at a postulated ligand entry/exit site in the α-subunit of hemoglobin, which, to the best of our knowledge, represents the first direct evidence for this site being used to access the heme iron. Hydrodisulfide, a postulated intermediate at the junction between thiosulfate and polysulfide formation, coordinates ferric hemoglobin and, in the presence of air, generated thiosulfate. At low sulfide/heme iron ratios, the product distribution between thiosulfate and iron-bound polysulfides was approximately equal. The iron-bound polysulfides were unstable at physiological glutathione concentrations and were reduced with concomitant formation of glutathione persulfide, glutathione disulfide, and H 2 S. Hence, although polysulfides are unlikely to be stable in the reducing intracellular milieu, glutathione persulfide could serve as a persulfide donor for protein persulfidation, a posttranslational modification by which H 2 S is postulated to signal.
- From the Department of Biological Chemistry, University of Michigan Medical School, Ann Arbor, Michigan 48109 and.
Organizational Affiliation: