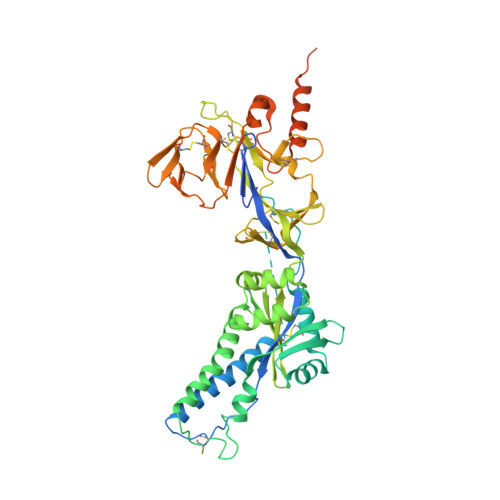
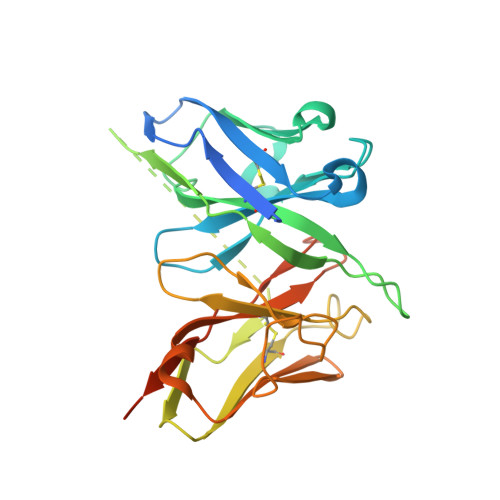
Structural basis for antibody cross-neutralization of respiratory syncytial virus and human metapneumovirus.
Wen, X., Mousa, J.J., Bates, J.T., Lamb, R.A., Crowe, J.E., Jardetzky, T.S.(2017) Nat Microbiol 2: 16272-16272
- PubMed: 28134915
- DOI: https://doi.org/10.1038/nmicrobiol.2016.272
- Primary Citation of Related Structures:
5U68 - PubMed Abstract:
Respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) are two closely related viruses that cause bronchiolitis and pneumonia in infants and the elderly 1 , with a significant health burden 2-6 . There are no licensed vaccines or small-molecule antiviral treatments specific to these two viruses at present. A humanized murine monoclonal antibody (palivizumab) is approved to treat high-risk infants for RSV infection 7,8 , but other treatments, as well as vaccines, for both viruses are still in development. Recent epidemiological modelling suggests that cross-immunity between RSV, HMPV and human parainfluenzaviruses may contribute to their periodic outbreaks 9 , suggesting that a deeper understanding of host immunity to these viruses may lead to enhanced strategies for their control. Cross-reactive neutralizing antibodies to the RSV and HMPV fusion (F) proteins have been identified 10,11 . Here, we examine the structural basis for cross-reactive antibody binding to RSV and HMPV F protein by two related, independently isolated antibodies, MPE8 and 25P13. We solved the structure of the MPE8 antibody bound to RSV F protein and identified the 25P13 antibody from an independent blood donor. Our results indicate that both antibodies use germline residues to interact with a conserved surface on F protein that could guide the emergence of cross-reactivity. The induction of similar cross-reactive neutralizing antibodies using structural vaccinology approaches could enhance intrinsic cross-immunity to these paramyxoviruses and approaches to controlling recurring outbreaks.
- Department of Structural Biology, Stanford University School of Medicine, Stanford, California 94305, USA.
Organizational Affiliation: