Double-flow focused liquid injector for efficient serial femtosecond crystallography.
Oberthuer, D., Knoska, J., Wiedorn, M.O., Beyerlein, K.R., Bushnell, D.A., Kovaleva, E.G., Heymann, M., Gumprecht, L., Kirian, R.A., Barty, A., Mariani, V., Tolstikova, A., Adriano, L., Awel, S., Barthelmess, M., Dorner, K., Xavier, P.L., Yefanov, O., James, D.R., Nelson, G., Wang, D., Calvey, G., Chen, Y., Schmidt, A., Szczepek, M., Frielingsdorf, S., Lenz, O., Snell, E., Robinson, P.J., Sarler, B., Belsak, G., Macek, M., Wilde, F., Aquila, A., Boutet, S., Liang, M., Hunter, M.S., Scheerer, P., Lipscomb, J.D., Weierstall, U., Kornberg, R.D., Spence, J.C., Pollack, L., Chapman, H.N., Bajt, S.(2017) Sci Rep 7: 44628-44628
- PubMed: 28300169
- DOI: https://doi.org/10.1038/srep44628
- Primary Citation of Related Structures:
5MND, 5U5Q - PubMed Abstract:
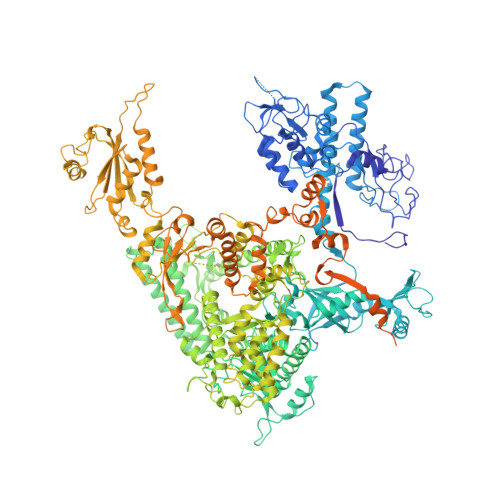
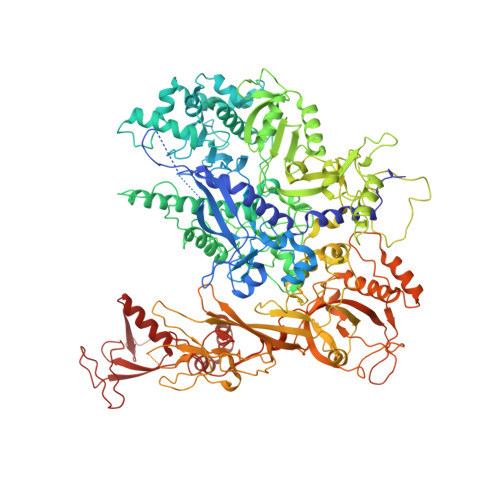
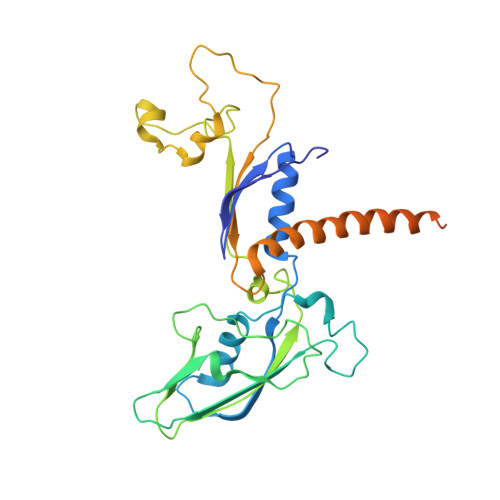
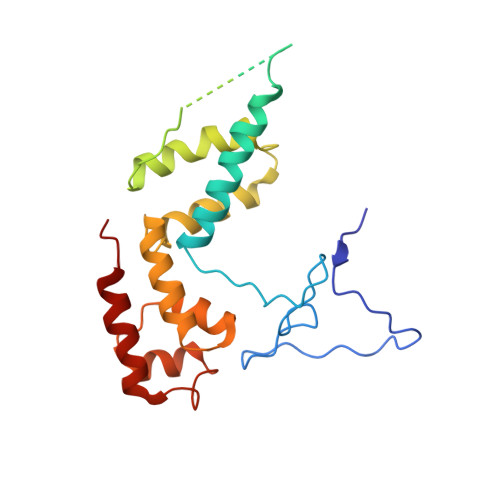
Serial femtosecond crystallography requires reliable and efficient delivery of fresh crystals across the beam of an X-ray free-electron laser over the course of an experiment. We introduce a double-flow focusing nozzle to meet this challenge, with significantly reduced sample consumption, while improving jet stability over previous generations of nozzles. We demonstrate its use to determine the first room-temperature structure of RNA polymerase II at high resolution, revealing new structural details. Moreover, the double-flow focusing nozzles were successfully tested with three other protein samples and the first room temperature structure of an extradiol ring-cleaving dioxygenase was solved by utilizing the improved operation and characteristics of these devices [corrected].
- Center for Free-Electron Laser Science, Deutsches Elektronen-Synchrotron DESY, Notkestraße 85, 22607 Hamburg, Germany.
Organizational Affiliation: