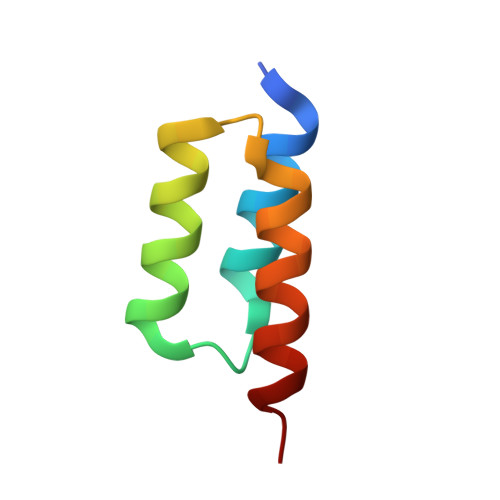
3-2-1: Structural insights from stepwise shrinkage of a three-helix Fc-binding domain to a single helix.
Ultsch, M., Braisted, A., Maun, H.R., Eigenbrot, C.(2017) Protein Eng Des Sel 30: 619-625
- PubMed: 28475752
- DOI: https://doi.org/10.1093/protein/gzx029
- Primary Citation of Related Structures:
5U4Y, 5U52, 5U66 - PubMed Abstract:
The well-studied B-domain from Staphylococcal protein A is a 59 amino acid three-helix bundle that binds the Fc portion of IgG with a dissociation constant of ~35 nM. The B-domain variant bearing a Gly to Ala mutation (=Z-domain) has been the subject of efforts to minimize a domain's size while retaining its function. We report X-ray crystallographic characterization of three steps in such a process using complexes with Fc: the full three-helix Z-domain, a 34 amino acid two-helix version called Z34C and a 13 amino acid single helix stabilized with an exo-helix tether, called LH1.
- Department of Structural Biology, Genentech Inc., 1 DNA Way, South San Francisco, CA 94080,USA.
Organizational Affiliation: