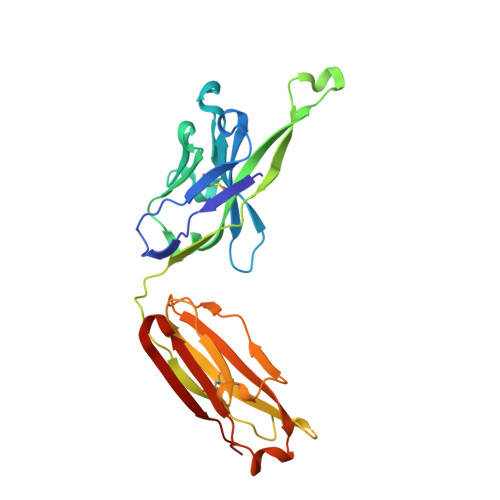
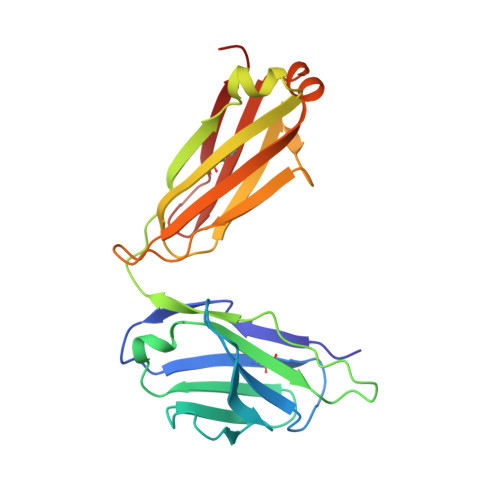
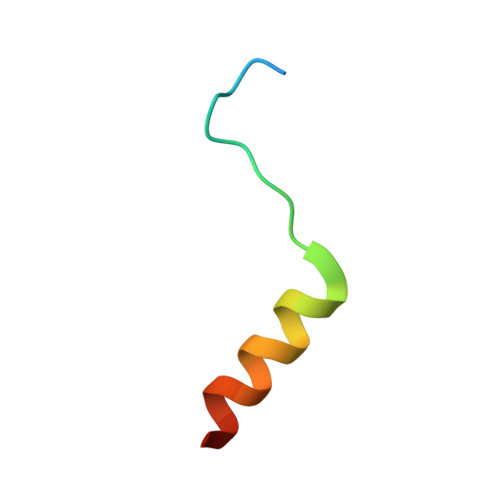
Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Williams, L.D., Ofek, G., Schatzle, S., McDaniel, J.R., Lu, X., Nicely, N.I., Wu, L., Lougheed, C.S., Bradley, T., Louder, M.K., McKee, K., Bailer, R.T., O'Dell, S., Georgiev, I.S., Seaman, M.S., Parks, R.J., Marshall, D.J., Anasti, K., Yang, G., Nie, X., Tumba, N.L., Wiehe, K., Wagh, K., Korber, B., Kepler, T.B., Munir Alam, S., Morris, L., Kamanga, G., Cohen, M.S., Bonsignori, M., Xia, S.M., Montefiori, D.C., Kelsoe, G., Gao, F., Mascola, J.R., Moody, M.A., Saunders, K.O., Liao, H.X., Tomaras, G.D., Georgiou, G., Haynes, B.F.(2017) Sci Immunol 2
- PubMed: 28783671
- DOI: https://doi.org/10.1126/sciimmunol.aal2200
- Primary Citation of Related Structures:
5U3J, 5U3K, 5U3L, 5U3M, 5U3N, 5U3O, 5U3P - PubMed Abstract:
Induction of broadly neutralizing antibodies (bnAbs) is a goal of HIV-1 vaccine development. Antibody 10E8, reactive with the distal portion of the membrane-proximal external region (MPER) of HIV-1 gp41, is broadly neutralizing. However, the ontogeny of distal MPER antibodies and the relationship of memory B cell to plasma bnAbs are poorly understood. HIV-1-specific memory B cell flow sorting and proteomic identification of anti-MPER plasma antibodies from an HIV-1-infected individual were used to isolate broadly neutralizing distal MPER bnAbs of the same B cell clonal lineage. Structural analysis demonstrated that antibodies from memory B cells and plasma recognized the envelope gp41 bnAb epitope in a distinct orientation compared with other distal MPER bnAbs. The unmutated common ancestor of this distal MPER bnAb was autoreactive, suggesting lineage immune tolerance control. Construction of chimeric antibodies of memory B cell and plasma antibodies yielded a bnAb that potently neutralized most HIV-1 strains.
- Duke Human Vaccine Institute, Duke University School of Medicine, Durham, NC 27710, USA.
Organizational Affiliation: