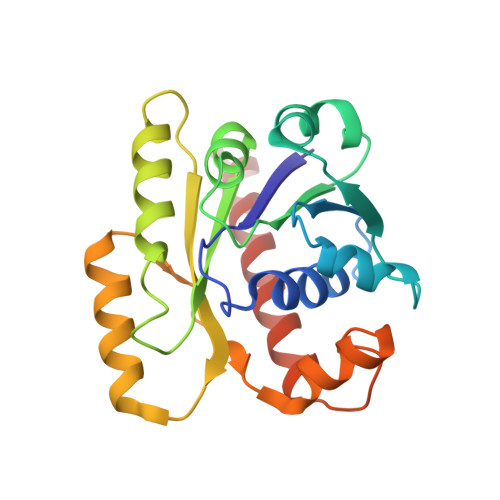
Structures of partition protein ParA with nonspecific DNA and ParB effector reveal molecular insights into principles governing Walker-box DNA segregation.
Zhang, H., Schumacher, M.A.(2017) Genes Dev 31: 481-492
- PubMed: 28373206
- DOI: https://doi.org/10.1101/gad.296319.117
- Primary Citation of Related Structures:
5U1G, 5U1J - PubMed Abstract:
Walker-box partition systems are ubiquitous in nature and mediate the segregation of bacterial and archaeal DNA. Well-studied plasmid Walker-box partition modules require ParA, centromere-DNA, and a centromere-binding protein, ParB. In these systems, ParA-ATP binds nucleoid DNA and uses it as a substratum to deliver ParB-attached cargo DNA, and ParB drives ParA dynamics, allowing ParA progression along the nucleoid. How ParA-ATP binds nonspecific DNA and is regulated by ParB is unclear. Also under debate is whether ParA polymerizes on DNA to mediate segregation. Here we describe structures of key ParA segregation complexes. The ParA-β,γ-imidoadenosine 5'-triphosphate (AMPPNP)-DNA structure revealed no polymers. Instead, ParA-AMPPNP dimerization creates a multifaceted DNA-binding surface, allowing it to preferentially bind high-density DNA regions (HDRs). DNA-bound ParA-AMPPNP adopts a dimer conformation distinct from the ATP sandwich dimer, optimized for DNA association. Our ParA-AMPPNP-ParB structure reveals that ParB binds at the ParA dimer interface, stabilizing the ATPase-competent ATP sandwich dimer, ultimately driving ParA DNA dissociation. Thus, the data indicate how harnessing a conformationally adaptive dimer can drive large-scale cargo movement without the requirement for polymers and suggest a segregation mechanism by which ParA-ATP dimers equilibrate to HDRs shown to be localized near cell poles of dividing chromosomes, thus mediating equipartition of attached ParB-DNA substrates.
- Department of Biochemistry, Duke University Medical Center, Durham, North Carolina 27710, USA.
Organizational Affiliation: