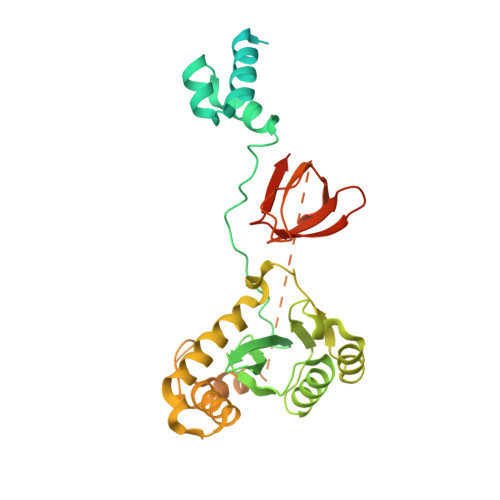
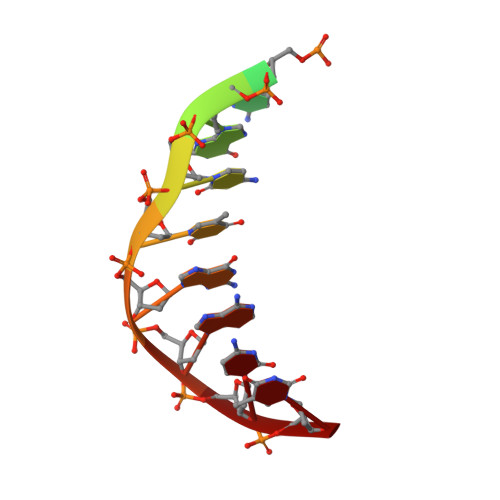
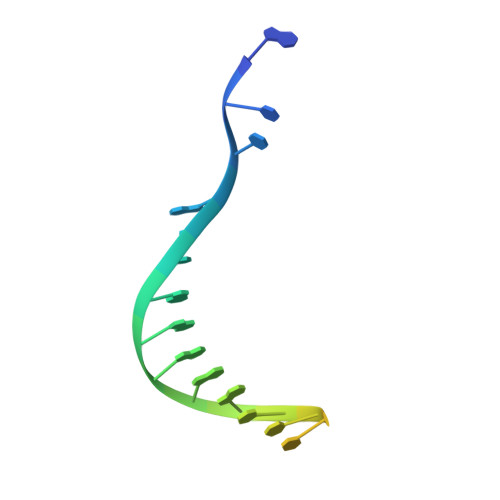
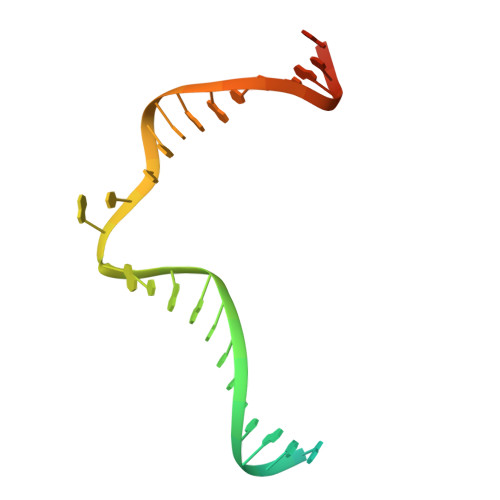
Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome.
Passos, D.O., Li, M., Yang, R., Rebensburg, S.V., Ghirlando, R., Jeon, Y., Shkriabai, N., Kvaratskhelia, M., Craigie, R., Lyumkis, D.(2017) Science 355: 89-92
- PubMed: 28059769
- DOI: https://doi.org/10.1126/science.aah5163
- Primary Citation of Related Structures:
5U1C - PubMed Abstract:
Like all retroviruses, HIV-1 irreversibly inserts a viral DNA (vDNA) copy of its RNA genome into host target DNA (tDNA). The intasome, a higher-order nucleoprotein complex composed of viral integrase (IN) and the ends of linear vDNA, mediates integration. Productive integration into host chromatin results in the formation of the strand transfer complex (STC) containing catalytically joined vDNA and tDNA. HIV-1 intasomes have been refractory to high-resolution structural studies. We used a soluble IN fusion protein to facilitate structural studies, through which we present a high-resolution cryo-electron microscopy (cryo-EM) structure of the core tetrameric HIV-1 STC and a higher-order form that adopts carboxyl-terminal domain rearrangements. The distinct STC structures highlight how HIV-1 can use the common retroviral intasome core architecture to accommodate different IN domain modules for assembly.
- Laboratory of Genetics and Helmsley Center for Genomic Medicine, The Salk Institute for Biological Studies, 10010 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: