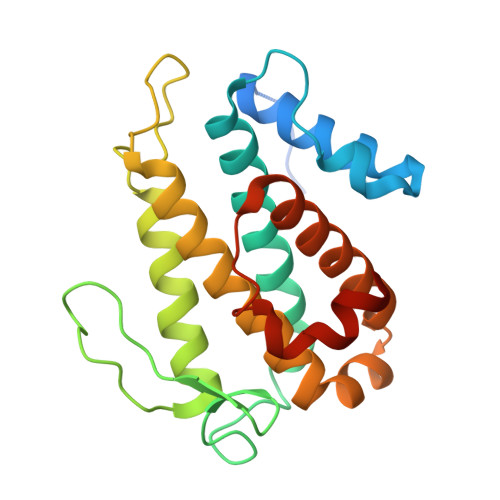
MOB1 Mediated Phospho-recognition in the Core Mammalian Hippo Pathway.
Couzens, A.L., Xiong, S., Knight, J.D.R., Mao, D.Y., Guettler, S., Picaud, S., Kurinov, I., Filippakopoulos, P., Sicheri, F., Gingras, A.C.(2017) Mol Cell Proteomics 16: 1098-1110
- PubMed: 28373298
- DOI: https://doi.org/10.1074/mcp.M116.065490
- Primary Citation of Related Structures:
5TWG, 5TWH - PubMed Abstract:
The Hippo tumor suppressor pathway regulates organ size and tissue homoeostasis in response to diverse signaling inputs. The core of the pathway consists of a short kinase cascade: MST1 and MST2 phosphorylate and activate LATS1 and LATS2, which in turn phosphorylate and inactivate key transcriptional coactivators, YAP1 and TAZ (gene WWTR1). The MOB1 adapter protein regulates both phosphorylation reactions firstly by concurrently binding to the upstream MST and downstream LATS kinases to enable the trans phosphorylation reaction, and secondly by allosterically activating the catalytic function of LATS1 and LATS2 to directly stimulate phosphorylation of YAP and TAZ. Studies of yeast Mob1 and human MOB1 revealed that the ability to recognize phosphopeptide sequences in their interactors, Nud1 and MST2 respectively, was critical to their roles in regulating the Mitotic Exit Network in yeast and the Hippo pathway in metazoans. However, the underlying rules of phosphopeptide recognition by human MOB1, the implications of binding specificity for Hippo pathway signaling, and the generality of phosphopeptide binding function to other human MOB family members remained elusive.Employing proteomics, peptide arrays and biochemical analyses, we systematically examine the phosphopeptide binding specificity of MOB1 and find it to be highly complementary to the substrate phosphorylation specificity of MST1 and MST2. We demonstrate that autophosphorylation of MST1 and MST2 on several threonine residues provides multiple MOB1 binding sites with varying binding affinities which in turn contribute to a redundancy of MST1-MOB1 protein interactions in cells. The crystal structures of MOB1A in complex with two favored phosphopeptide sites in MST1 allow for a full description of the MOB1A phosphopeptide-binding consensus. Lastly, we show that the phosphopeptide binding properties of MOB1A are conserved in all but one of the seven MOB family members in humans, thus providing a starting point for uncovering their elusive cellular functions.
- From the ‡Lunenfeld-Tanenbaum Research Institute, Sinai Health System, Toronto, Ontario, Canada, M5G 1X5.
Organizational Affiliation: