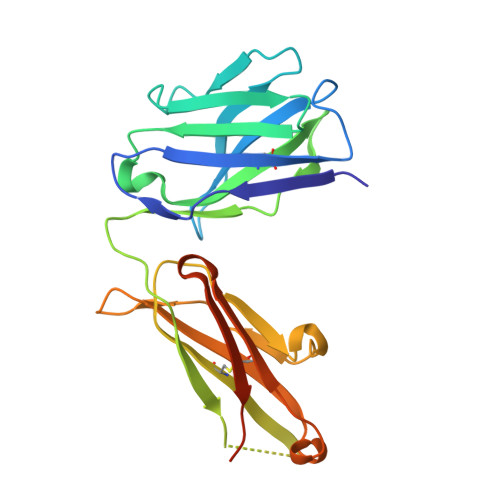
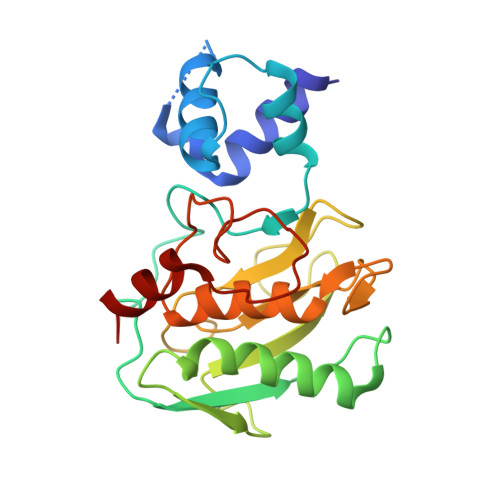
Biochemical characterization and structure determination of a potent, selective antibody inhibitor of human MMP9.
Appleby, T.C., Greenstein, A.E., Hung, M., Liclican, A., Velasquez, M., Villasenor, A.G., Wang, R., Wong, M.H., Liu, X., Papalia, G.A., Schultz, B.E., Sakowicz, R., Smith, V., Kwon, H.J.(2017) J Biological Chem 292: 6810-6820
- PubMed: 28235803
- DOI: https://doi.org/10.1074/jbc.M116.760579
- Primary Citation of Related Structures:
5TH6, 5TH9 - PubMed Abstract:
Matrix metalloproteinase 9 (MMP9) is a member of a large family of proteases that are secreted as inactive zymogens. It is a key regulator of the extracellular matrix, involved in the degradation of various extracellular matrix proteins. MMP9 plays a pathological role in a variety of inflammatory and oncology disorders and has long been considered an attractive therapeutic target. GS-5745, a potent, highly selective humanized monoclonal antibody inhibitor of MMP9, has shown promise in treating ulcerative colitis and gastric cancer. Here we describe the crystal structure of GS-5745·MMP9 complex and biochemical studies to elucidate the mechanism of inhibition of MMP9 by GS-5745. GS-5745 binds MMP9 distal to the active site, near the junction between the prodomain and catalytic domain, and inhibits MMP9 by two mechanisms. Binding to pro-MMP9 prevents MMP9 activation, whereas binding to active MMP9 allosterically inhibits activity.
- From Gilead Sciences, Inc., Foster City, California 94404.
Organizational Affiliation: