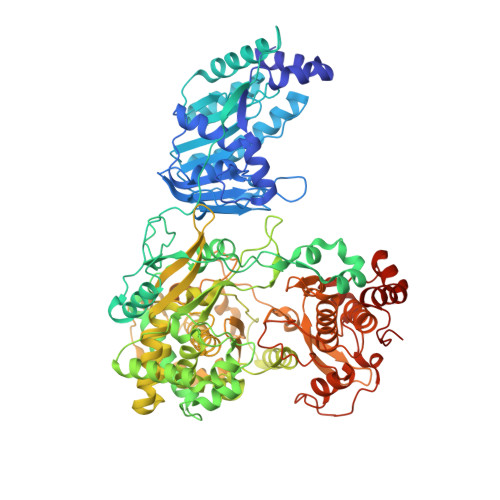
Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5.
Upadhyay, A.K., Cyr, M., Longenecker, K., Tripathi, R., Sun, C., Kempf, D.J.(2017) Acta Crystallogr F Struct Biol Commun 73: 116-122
- PubMed: 28291746
- DOI: https://doi.org/10.1107/S2053230X17001601
- Primary Citation of Related Structures:
5TFR - PubMed Abstract:
The rapid spread of the recent Zika virus (ZIKV) epidemic across various countries in the American continent poses a major health hazard for the unborn fetuses of pregnant women. To date, there is no effective medical intervention. The nonstructural protein 5 of Zika virus (ZIKV-NS5) is critical for ZIKV replication through the 5'-RNA capping and RNA polymerase activities present in its N-terminal methyltransferase (MTase) and C-terminal RNA-dependent RNA polymerase (RdRp) domains, respectively. The crystal structure of the full-length ZIKV-NS5 protein has been determined at 3.05 Å resolution from a crystal belonging to space group P2 1 2 1 2 and containing two protein molecules in the asymmetric unit. The structure is similar to that reported for the NS5 protein from Japanese encephalitis virus and suggests opportunities for structure-based drug design targeting either its MTase or RdRp domain.
- AbbVie Inc., 1 North Waukegan Road, North Chicago, IL 60064, USA.
Organizational Affiliation: