Structural basis for clonal diversity of the human T-cell response to a dominant influenza virus epitope.
Yang, X., Chen, G., Weng, N.P., Mariuzza, R.A.(2017) J Biological Chem 292: 18618-18627
- PubMed: 28931605
- DOI: https://doi.org/10.1074/jbc.M117.810382
- Primary Citation of Related Structures:
5TEZ - PubMed Abstract:
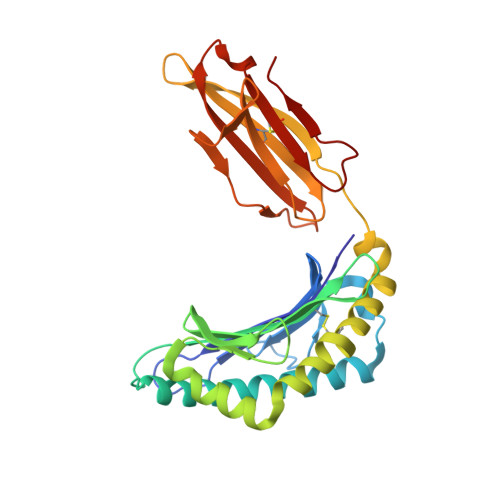
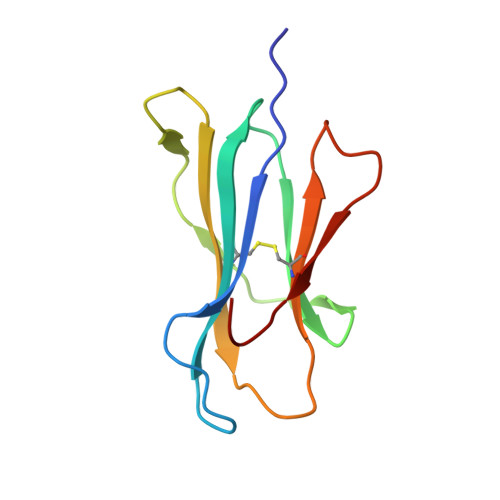
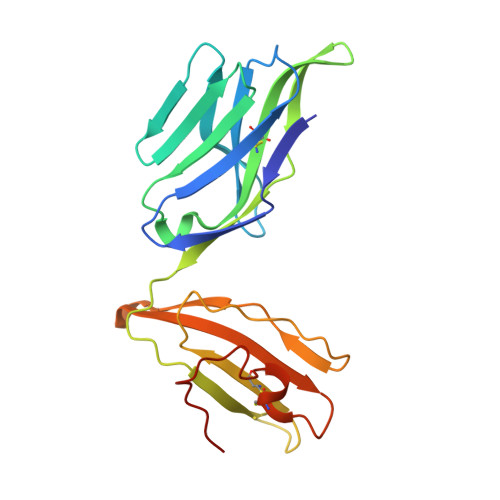
Influenza A virus (IAV) causes an acute infection in humans that is normally eliminated by CD8 + cytotoxic T lymphocytes. Individuals expressing the MHC class I molecule HLA-A2 produce cytotoxic T lymphocytes bearing T-cell receptors (TCRs) that recognize the immunodominant IAV epitope GILGFVFTL (GIL). Most GIL-specific TCRs utilize α/β chain pairs encoded by the TRAV27/TRBV19 gene combination to recognize this relatively featureless peptide epitope (canonical TCRs). However, ∼40% of GIL-specific TCRs express a wide variety of other TRAV/TRBV combinations (non-canonical TCRs). To investigate the structural underpinnings of this remarkable diversity, we determined the crystal structure of a non-canonical GIL-specific TCR (F50) expressing the TRAV13-1/TRBV27 gene combination bound to GIL-HLA-A2 to 1.7 Å resolution. Comparison of the F50-GIL-HLA-A2 complex with the previously published complex formed by a canonical TCR (JM22) revealed that F50 and JM22 engage GIL-HLA-A2 in markedly different orientations. These orientations are distinguished by crossing angles of TCR to peptide-MHC of 29° for F50 versus 69° for JM22 and by a focus by F50 on the C terminus rather than the center of the MHC α1 helix for JM22. In addition, F50, unlike JM22, uses a tryptophan instead of an arginine to fill a critical notch between GIL and the HLA-A2 α2 helix. The F50-GIL-HLA-A2 complex shows that there are multiple structurally distinct solutions to recognizing an identical peptide-MHC ligand with sufficient affinity to elicit a broad anti-IAV response that protects against viral escape and T-cell clonal loss.
- From the University of Maryland Institute for Bioscience and Biotechnology Research, W. M. Keck Laboratory for Structural Biology, Rockville, Maryland 20850.
Organizational Affiliation: