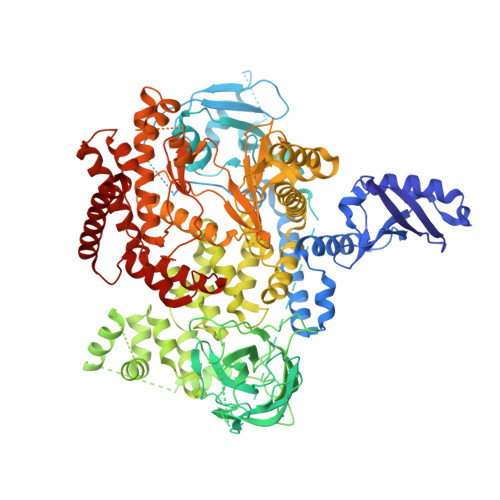
Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
Castanedo, G.M., Blaquiere, N., Beresini, M., Bravo, B., Brightbill, H., Chen, J., Cui, H.F., Eigenbrot, C., Everett, C., Feng, J., Godemann, R., Gogol, E., Hymowitz, S., Johnson, A., Kayagaki, N., Kohli, P.B., Knuppel, K., Kraemer, J., Kruger, S., Loke, P., McEwan, P., Montalbetti, C., Roberts, D.A., Smith, M., Steinbacher, S., Sujatha-Bhaskar, S., Takahashi, R., Wang, X., Wu, L.C., Zhang, Y., Staben, S.T.(2017) J Med Chem 60: 627-640
- PubMed: 28005357
- DOI: https://doi.org/10.1021/acs.jmedchem.6b01363
- Primary Citation of Related Structures:
5T8F, 5T8O, 5T8P, 5T8Q - PubMed Abstract:
We report here structure-guided optimization of a novel series of NF-κB inducing kinase (NIK) inhibitors. Starting from a modestly potent, low molecular weight lead, activity was improved by designing a type 11/2 binding mode that accessed a back pocket past the methionine-471 gatekeeper. Divergent binding modes in NIK and PI3K were exploited to dampen PI3K inhibition while maintaining NIK inhibition within these series. Potent compounds were discovered that selectively inhibit the nuclear translocation of NF-κB2 (p52/REL-B) but not canonical NF-κB1 (REL-A/p50).
- Genentech, Inc. 1 DNA Way, South San Francisco, California 94080, United States.
Organizational Affiliation: