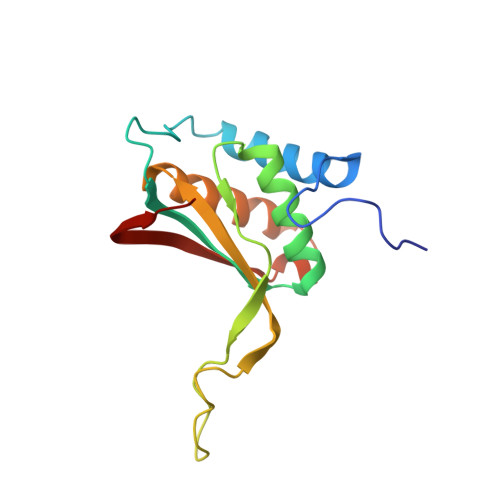
Carcinoma-risk variant of EBNA1 deregulates Epstein-Barr Virus episomal latency.
Dheekollu, J., Malecka, K., Wiedmer, A., Delecluse, H.J., Chiang, A.K., Altieri, D.C., Messick, T.E., Lieberman, P.M.(2017) Oncotarget 8: 7248-7264
- PubMed: 28077791
- DOI: https://doi.org/10.18632/oncotarget.14540
- Primary Citation of Related Structures:
5T7X - PubMed Abstract:
Epstein-Barr Virus (EBV) latent infection is a causative co-factor for endemic Nasopharyngeal Carcinoma (NPC). NPC-associated variants have been identified in EBV-encoded nuclear antigen EBNA1. Here, we solve the X-ray crystal structure of an NPC-derived EBNA1 DNA binding domain (DBD) and show that variant amino acids are found on the surface away from the DNA binding interface. We show that NPC-derived EBNA1 is compromised for DNA replication and episome maintenance functions. Recombinant virus containing the NPC EBNA1 DBD are impaired in their ability to immortalize primary B-lymphocytes and suppress lytic transcription during early stages of B-cell infection. We identify Survivin as a host protein deficiently bound by the NPC variant of EBNA1 and show that Survivin depletion compromises EBV episome maintenance in multiple cell types. We propose that endemic variants of EBNA1 play a significant role in EBV-driven carcinogenesis by altering key regulatory interactions that destabilize latent infection.
- The Wistar Institute, Philadelphia, PA USA.
Organizational Affiliation: