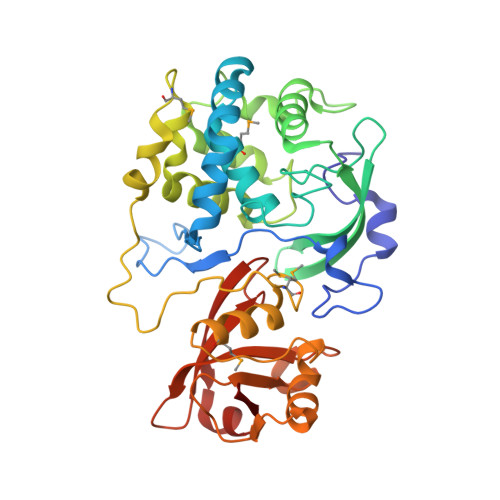
2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus.
Minasov, G., Nocadello, S., Shuvalova, L., Kiryukhina, O., Dubrovska, I., Bagnoli, F., Grandi, G., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.