Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Bzymek, K.P., Avery, K.A., Ma, Y., Horne, D.A., Williams, J.C.(2016) Acta Crystallogr F Struct Biol Commun 72: 820-830
- PubMed: 27834791
- DOI: https://doi.org/10.1107/S2053230X16016149
- Primary Citation of Related Structures:
5ETU, 5EUK, 5F88, 5FF6, 5I2I, 5IOP, 5IR1, 5ITF, 5IV2, 5IVZ, 5T1K, 5T1L, 5T1M, 5TH2 - PubMed Abstract:
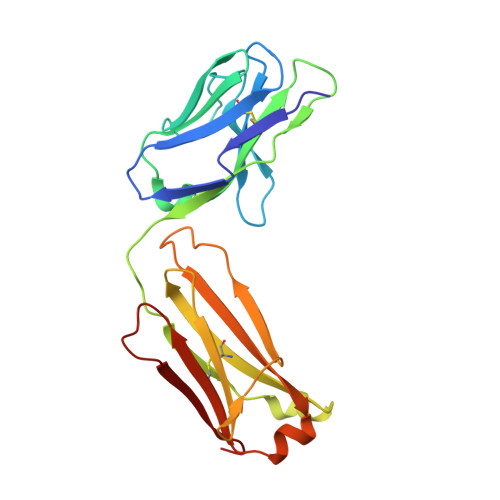
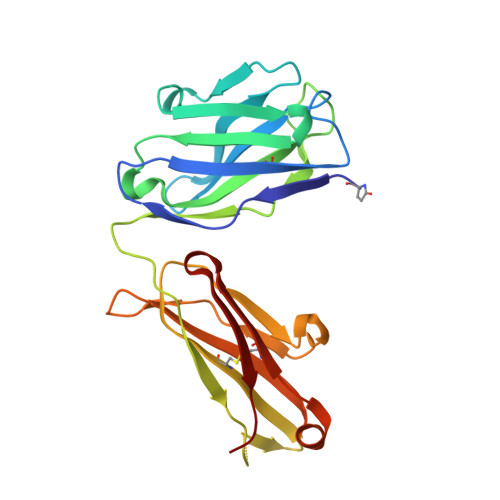
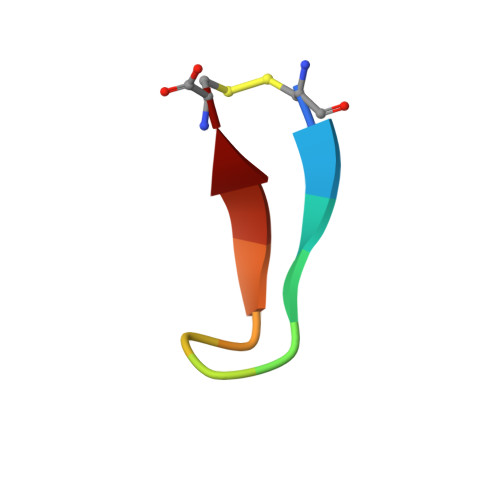
Herein, multiple crystal structures of meditope peptide derivatives incorporating natural and unnatural amino acids bound to the cetuximab Fab domain are presented. The affinity of each derivative was determined by surface plasmon resonance and correlated to the atomic structure. Overall, it was observed that the hydrophobic residues in the meditope peptide, Phe3, Leu5 and Leu10, could accommodate a number of moderate substitutions, but these invariably reduced the overall affinity and half-life of the interaction. In one case, the substitution of Phe3 by histidine led to a change in the rotamer conformation, in which the imidazole ring flipped to a solvent-exposed position. Based on this observation, Phe3 was substituted by diphenylalanine and it was found that the phenyl rings in this variant mimic the superposition of the Phe3 and His3 structures, producing a moderate increase, of 1.4-fold, in the half-life of the complex. In addition, it was observed that substitution of Leu5 by tyrosine and glutamate strongly reduced the affinity, whereas the substitution of Leu5 by diphenylalanine moderately reduced the half-life (by approximately fivefold). Finally, it was observed that substitution of Arg8 and Arg9 by citrulline dramatically reduced the overall affinity, presumably owing to lost electrostatic interactions. Taken together, these studies provide insight into the meditope-cetuximab interaction at the atomic level.
- Department of Molecular Medicine, Beckman Research Institute of City of Hope, 1710 Flower Street, Duarte, CA 91010, USA.
Organizational Affiliation: