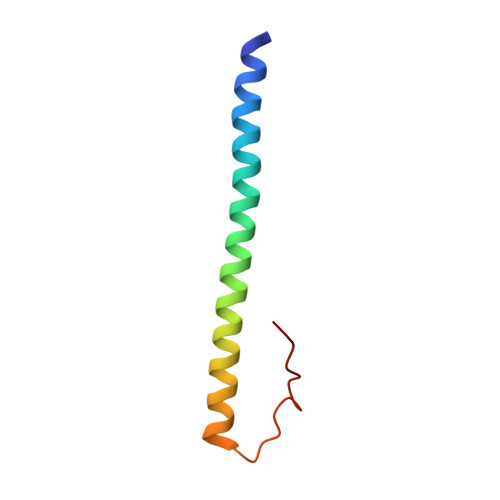
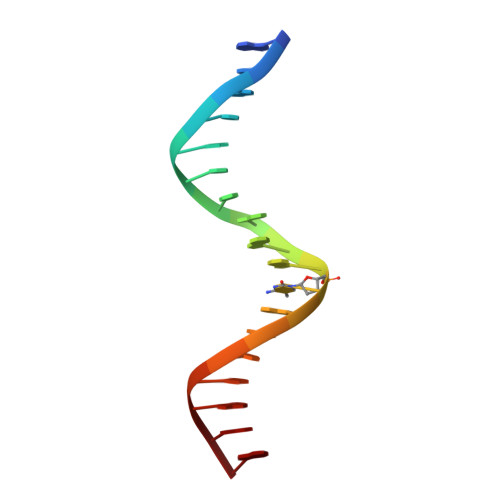
Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Hong, S., Wang, D., Horton, J.R., Zhang, X., Speck, S.H., Blumenthal, R.M., Cheng, X.(2017) Nucleic Acids Res 45: 2503-2515
- PubMed: 28158710
- DOI: https://doi.org/10.1093/nar/gkx057
- Primary Citation of Related Structures:
5SZX, 5T01 - PubMed Abstract:
Activator protein 1 (AP-1) is a transcription factor that recognizes two versions of a 7-base pair response element, either 5΄- GAG CA-3΄ or 5΄- GAG CA-3΄ (where M = 5-methylcytosine). These two elements share the feature that 5-methylcytosine and thymine both have a methyl group in the same position, 5-carbon of the pyrimidine, so each of them has two methyl groups at nucleotide positions 1 and 5 from the 5΄ end, resulting in four methyl groups symmetrically positioned in duplex DNA. Epstein-Barr Virus Zta is a key transcriptional regulator of the viral lytic cycle that is homologous to AP-1. Zta recognizes several methylated Zta-response elements, including meZRE1 (5΄- GAG C A-3΄) and meZRE2 (5΄- GAG G A-3΄), where a methylated cytosine occupies one of the inner thymine residues corresponding to the AP-1 element, resulting in the four spatially equivalent methyl groups. Here, we study how AP-1 and Zta recognize these methyl groups within their cognate response elements. These methyl groups are in van der Waals contact with a conserved di-alanine in AP-1 dimer (Ala265 and Ala266 in Jun), or with the corresponding Zta residues Ala185 and Ser186 (via its side chain carbon Cβ atom). Furthermore, the two ZRE elements differ at base pair 6 (C:G versus G:C), forming a pseudo-symmetric sequence (meZRE1) or an asymmetric sequence (meZRE2). In vitro DNA binding assays suggest that Zta has high affinity for all four sequences examined, whereas AP-1 has considerably reduced affinity for the asymmetric sequence (meZRE2). We ascribe this difference to Zta Ser186 (a unique residue for Zta) whose side chain hydroxyl oxygen atom interacts with the two half sites differently, whereas the corresponding Ala266 of AP-1 Jun protein lacks such flexibility. Our analyses demonstrate a novel mechanism of 5mC/T recognition in a methylation-dependent, spatial and sequence-specific approach by basic leucine-zipper transcriptional factors.
- Department of Biochemistry, Emory University School of Medicine, 1510 Clifton Road, Atlanta, GA 30322, USA.
Organizational Affiliation: