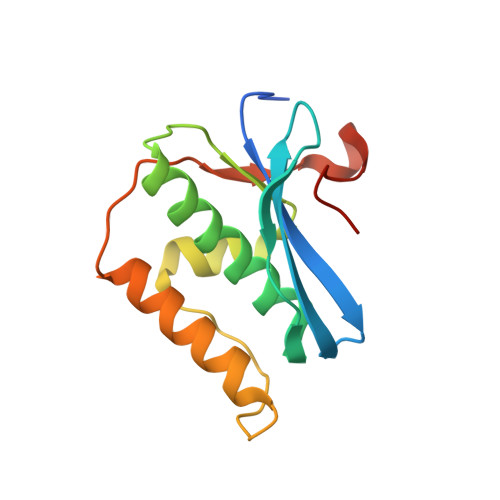
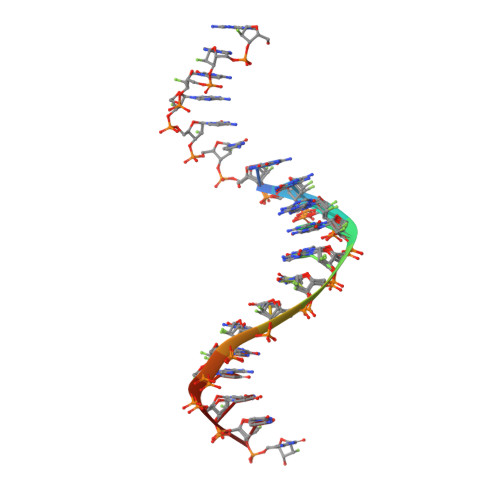
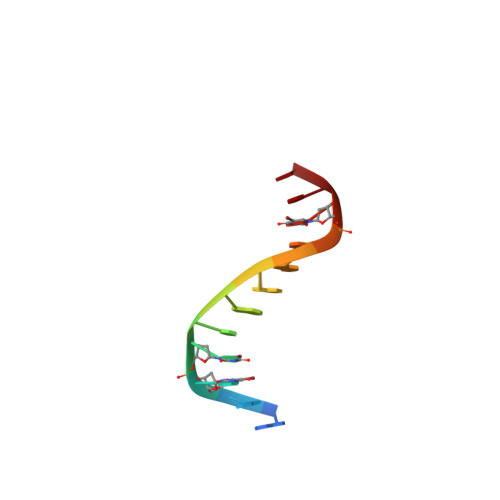
Limits of RNA 2'-OH Mimicry by Fluorine: Crystal Structure of Bacillus halodurans RNase H Bound to a 2'-FRNA:DNA Hybrid.
Pallan, P.S., Prakash, T.P., de Leon, A.R., Egli, M.(2016) Biochemistry 55: 5321-5325
- PubMed: 27611889
- DOI: https://doi.org/10.1021/acs.biochem.6b00849
- Primary Citation of Related Structures:
5SWM - PubMed Abstract:
RNase H1 cleaves the RNA strand of RNA:DNA hybrids. Replacement of RNA 2'-hydroxyls by fluorine (FRNA) is commonly used to stabilize aptamers and siRNAs. However, FRNA:DNA hybrids fail to elicit RNase H activity. The underlying reasons are unclear, as 2'-OH groups are not directly involved in cleavage. We determined the crystal structure of Bacillus halodurans RNase H bound to a FRNA:DNA hybrid. The structure points to dynamic (slippage of the FRNA:DNA hybrid relative to the enzyme), geometric (different curvatures of FRNA:DNA and RNA:DNA hybrids), and electronic reasons (Mg(2+) absent from the active site of the FRNA:DNA complex) for the loss of RNaseH activity.
- Department of Biochemistry, Vanderbilt University, School of Medicine , Nashville, Tennessee 37232, United States.
Organizational Affiliation: