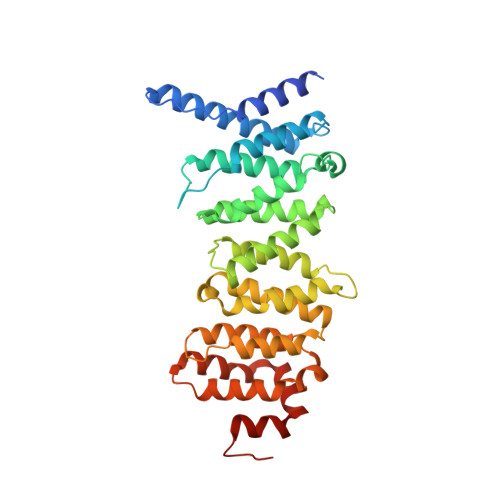
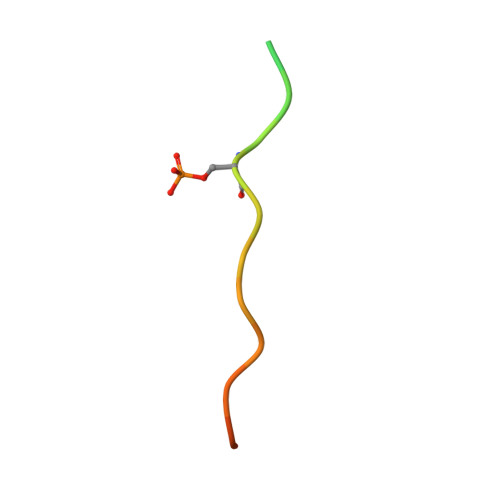
Expanding the PP2A Interactome by Defining a B56-Specific SLiM.
Wang, X., Bajaj, R., Bollen, M., Peti, W., Page, R.(2016) Structure 24: 2174-2181
- PubMed: 27998540
- DOI: https://doi.org/10.1016/j.str.2016.09.010
- Primary Citation of Related Structures:
5K6S, 5SW9, 5SWF - PubMed Abstract:
Specific interactions between proteins govern essential physiological processes including signaling. Many enzymes, especially the family of serine/threonine phosphatases (PSPs: PP1, PP2A, and PP2B/calcineurin/CN), recruit substrates and regulatory proteins by binding short linear motifs (SLiMs), short sequences found within intrinsically disordered regions that mediate specific protein-protein interactions. While tremendous progress had been made in identifying where and how SLiMs bind PSPs, especially PP1 and CN, essentially nothing is known about how SLiMs bind PP2A, a validated cancer drug target. Here we describe three structures of a PP2A-SLiM interaction (B56:pS-RepoMan, B56:pS-BubR1, and B56:pSpS-BubR1), show that this PP2A-specific SLiM is defined as LSPIxE, and then use these data to discover scores of likely PP2A regulators and substrates. Together, these data provide a powerful approach not only for dissecting PP2A interaction networks in cells but also for targeting PP2A diseases, such as cancer.
- Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, RI 02912, USA.
Organizational Affiliation: