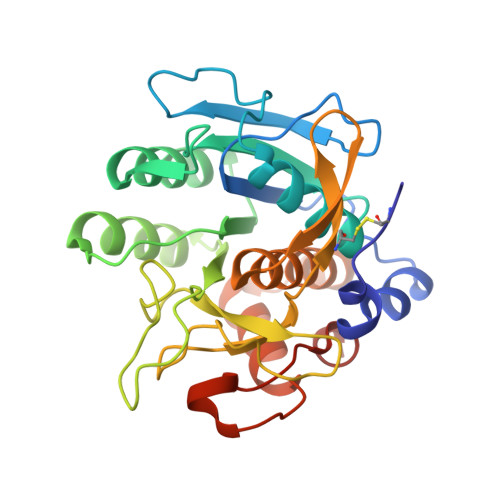
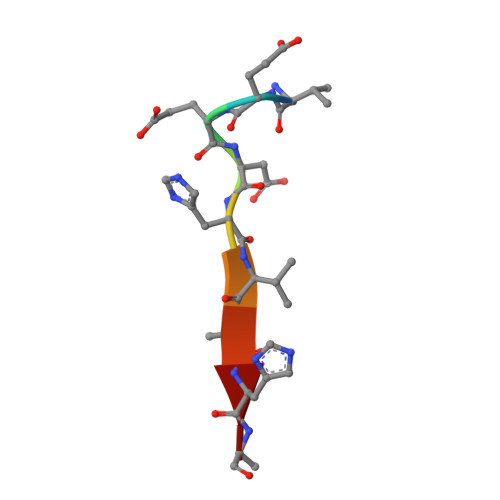
Design of a substrate-tailored peptiligase variant for the efficient synthesis of thymosin-alpha1.
Schmidt, M., Toplak, A., Rozeboom, H.J., Wijma, H.J., Quaedflieg, P.J.L.M., van Maarseveen, J.H., Janssen, D.B., Nuijens, T.(2018) Org Biomol Chem 16: 609-618
- PubMed: 29300408
- DOI: https://doi.org/10.1039/c7ob02812a
- Primary Citation of Related Structures:
5OX2 - PubMed Abstract:
The synthesis of thymosin-α 1 , an acetylated 28 amino acid long therapeutic peptide, via conventional chemical methods is exceptionally challenging. The enzymatic coupling of unprotected peptide segments in water offers great potential for a more efficient synthesis of peptides that are difficult to synthesize. Based on the design of a highly engineered peptide ligase, we developed a fully convergent chemo-enzymatic peptide synthesis (CEPS) process for the production of thymosin-α 1 via a 14-mer + 14-mer segment condensation strategy. Using structure-inspired enzyme engineering, the thiol-subtilisin variant peptiligase was tailored to recognize the respective 14-mer thymosin-α 1 segments in order to create a clearly improved biocatalyst, termed thymoligase. Thymoligase catalyzes peptide bond formation between both segments with a very high efficiency (>94% yield) and is expected to be well applicable to many other ligations in which residues with similar characteristics (e.g. Arg and Glu) are present in the respective positions P1 and P1'. The crystal structure of thymoligase was determined and shown to be in good agreement with the model used for the engineering studies. The combination of the solid phase peptide synthesis (SPPS) of the 14-mer segments and their thymoligase-catalyzed ligation on a gram scale resulted in a significantly increased, two-fold higher overall yield (55%) of thymosin-α 1 compared to those typical of existing industrial processes.
- EnzyPep B.V., Brightlands Campus, Urmonderbaan 22, 6167 RD Geleen, The Netherlands. timo@enzypep.com.
Organizational Affiliation: