Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-beta homologue, Kap95p.
Liu, S.M., Stewart, M.(2005) J Mol Biology 349: 515-525
- PubMed: 15878174
- DOI: https://doi.org/10.1016/j.jmb.2005.04.003
- Primary Citation of Related Structures:
5OWU - PubMed Abstract:
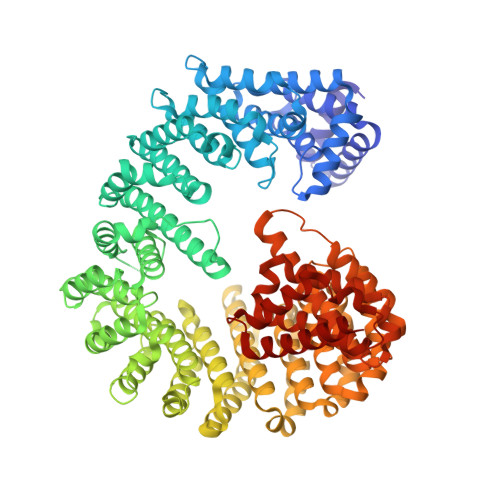
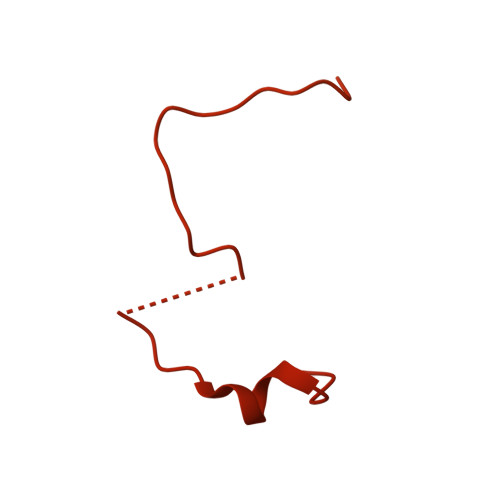
Macromolecules are transported across the nuclear envelope most frequently by karyopherin/importin-beta superfamily members that are constructed from HEAT repeats. Transport of Kap95p (yeast importin-beta), the principal carrier for protein import, through nuclear pore complexes is facilitated by interactions with nucleoporins containing FG repeats. However, Nup1p interacts more strongly with Kap95p than other FG-nucleoporins. To establish the basis of this increased affinity, we determined the structure of Kap95p complexed with Nup1p residues 963-1076 that contain the high-affinity Kap95p binding site. Nup1p binds Kap95p at three sites between the outer A-helices of HEAT repeats 5, 6, 7 and 8. At each site, phenylalanine residues from Nup1p are buried in hydrophobic depressions between adjacent HEAT repeats. Although the Nup1p and generic FG-nucleoporin binding sites on Kap95p overlap, Nup1p binding differs markedly and has contributions from additional hydrophobic residues, together with interactions generated by the intimate contact of the linker between Nup1 residues 977-987 with Kap95p. The length and composition of this linker is crucial and suggests how differences in affinity for Kap95p both between and within FG-nucleoporins arise.
- MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK.
Organizational Affiliation: