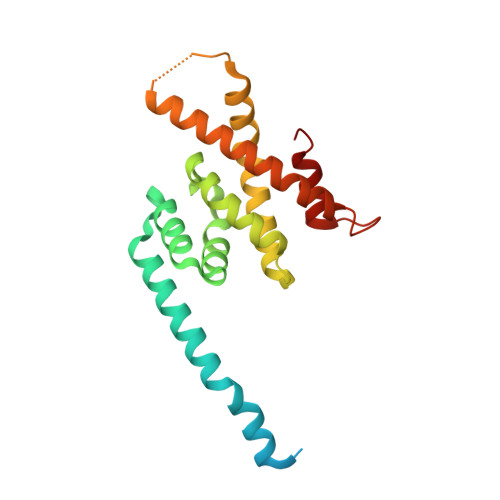
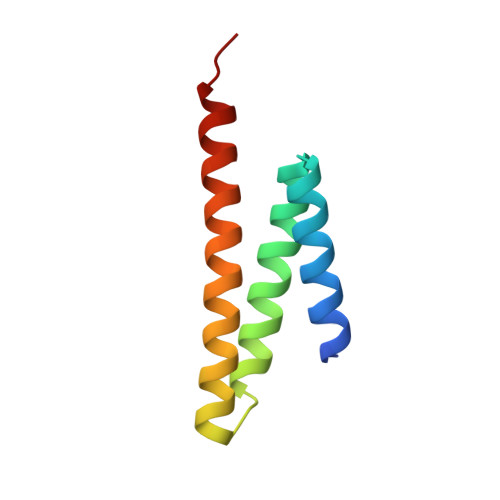
Structural Basis of Formation of the Microtubule Minus-End-Regulating CAMSAP-Katanin Complex.
Jiang, K., Faltova, L., Hua, S., Capitani, G., Prota, A.E., Landgraf, C., Volkmer, R., Kammerer, R.A., Steinmetz, M.O., Akhmanova, A.(2018) Structure 26: 375-382.e4
- PubMed: 29395789
- DOI: https://doi.org/10.1016/j.str.2017.12.017
- Primary Citation of Related Structures:
5OW5 - PubMed Abstract:
CAMSAP/Patronin family members regulate the organization and stability of microtubule minus ends in various systems ranging from mitotic spindles to differentiated epithelial cells and neurons. Mammalian CAMSAP2 and CAMSAP3 bind to growing microtubule minus ends, where they form stretches of stabilized microtubule lattice. The microtubule-severing ATPase katanin interacts with CAMSAPs and limits the length of CAMSAP-decorated microtubule stretches. Here, by using biochemical, biophysical, and structural approaches, we reveal that a short helical motif conserved in CAMSAP2 and CAMSAP3 binds to the heterodimer formed by the N- and C-terminal domains of katanin subunits p60 and p80, respectively. The identified CAMSAP-katanin binding mode is supported by mutational analysis and genome-editing experiments. It is strikingly similar to the one seen in the ASPM-katanin complex, which is responsible for microtubule minus-end regulation in mitotic spindles. Our work provides a general molecular mechanism for the cooperation of katanin with major microtubule minus-end regulators.
- Cell Biology, Department of Biology, Faculty of Science, Utrecht University, Padualaan 8, 3584 Utrecht, the Netherlands.
Organizational Affiliation: