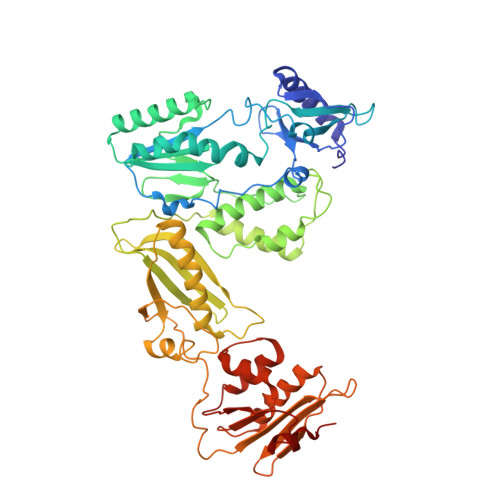
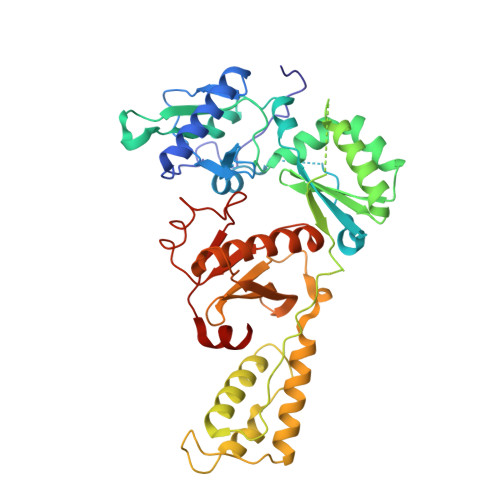
The structure of FIV reverse transcriptase and its implications for non-nucleoside inhibitor resistance.
Galilee, M., Alian, A.(2018) PLoS Pathog 14: e1006849-e1006849
- PubMed: 29364950
- DOI: https://doi.org/10.1371/journal.ppat.1006849
- Primary Citation of Related Structures:
5OVN - PubMed Abstract:
Reverse transcriptase (RT) is the target for the majority of anti-HIV-1 drugs. As with all anti-AIDS treatments, continued success of RT inhibitors is persistently disrupted by the occurrence of resistance mutations. To explore latent resistance mechanisms potentially accessible to therapeutically challenged HIV-1 viruses, we examined RT from the related feline immunodeficiency virus (FIV). FIV closely parallels HIV-1 in its replication and pathogenicity, however, is resistant to all non-nucleoside inhibitors (NNRTI). The intrinsic resistance of FIV RT is particularly interesting since FIV harbors the Y181 and Y188 sensitivity residues absent in both HIV-2 and SIV. Unlike RT from HIV-2 or SIV, previous efforts have failed to make FIV RT susceptible to NNRTIs concluding that the structure or flexibility of the feline enzyme must be profoundly different. We report the first crystal structure of FIV RT and, being the first structure of an RT from a non-primate lentivirus, enrich the structural and species repertoires available for RT. The structure demonstrates that while the NNRTI binding pocket is conserved, minor subtleties at the entryway can render the FIV RT pocket more restricted and unfavorable for effective NNRTI binding. Measuring NNRTI binding affinity to FIV RT shows that the "closed" pocket configuration inhibits NNRTI binding. Mutating the loop residues rimming the entryway of FIV RT pocket allows for NNRTI binding, however, it does not confer sensitivity to these inhibitors. This reveals a further layer of resistance caused by inherent FIV RT variances that could have enhanced the dissociation of bound inhibitors, or, perhaps, modulated protein plasticity to overcome inhibitory effects of bound NNRTIs. The more "closed" conformation of FIV RT pocket can provide a template for the development of innovative drugs that could unlock the constrained pocket, and the resilient mutant version of the enzyme can offer a fresh model for the study of NNRTI-resistance mechanisms overlooked in HIV-1.
- Faculty of Biology, Technion-Israel Institute of Technology, Haifa, Israel.
Organizational Affiliation: