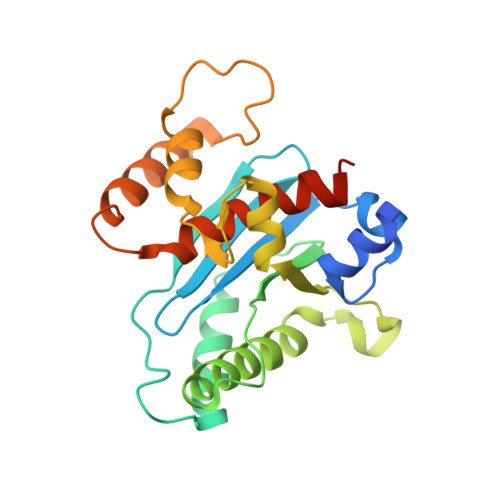
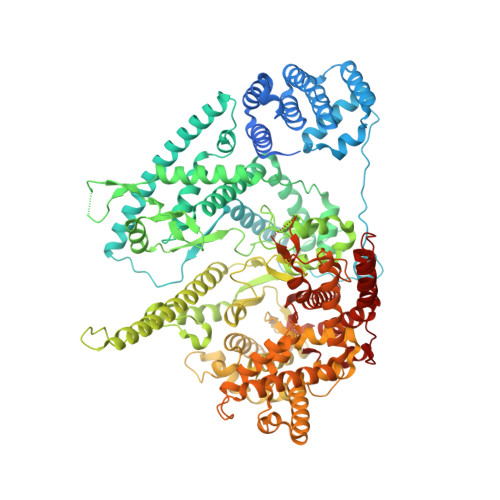
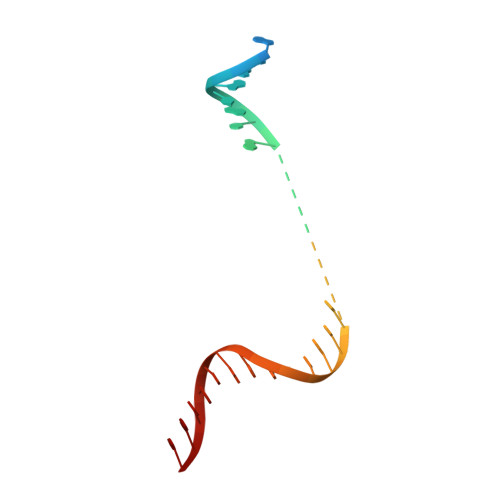
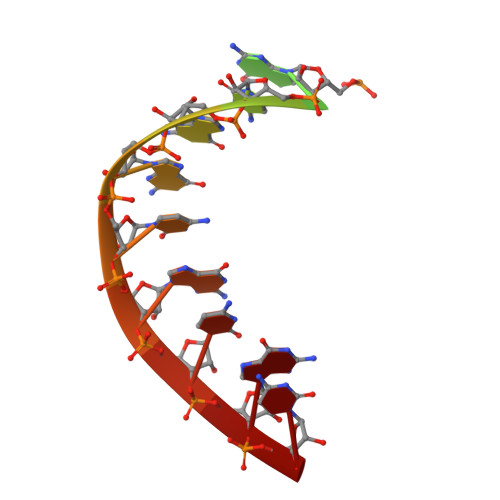
Mechanism of Transcription Anti-termination in Human Mitochondria.
Hillen, H.S., Parshin, A.V., Agaronyan, K., Morozov, Y.I., Graber, J.J., Chernev, A., Schwinghammer, K., Urlaub, H., Anikin, M., Cramer, P., Temiakov, D.(2017) Cell 171: 1082-1093.e13
- PubMed: 29033127
- DOI: https://doi.org/10.1016/j.cell.2017.09.035
- Primary Citation of Related Structures:
5OL8, 5OL9, 5OLA - PubMed Abstract:
In human mitochondria, transcription termination events at a G-quadruplex region near the replication origin are thought to drive replication of mtDNA by generation of an RNA primer. This process is suppressed by a key regulator of mtDNA-the transcription factor TEFM. We determined the structure of an anti-termination complex in which TEFM is bound to transcribing mtRNAP. The structure reveals interactions of the dimeric pseudonuclease core of TEFM with mobile structural elements in mtRNAP and the nucleic acid components of the elongation complex (EC). Binding of TEFM to the DNA forms a downstream "sliding clamp," providing high processivity to the EC. TEFM also binds near the RNA exit channel to prevent formation of the RNA G-quadruplex structure required for termination and thus synthesis of the replication primer. Our data provide insights into target specificity of TEFM and mechanisms by which it regulates the switch between transcription and replication of mtDNA.
- Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany.
Organizational Affiliation: