Structure-Activity Relationships and Biological Characterization of a Novel, Potent, and Serum Stable C-X-C Chemokine Receptor Type 4 (CXCR4) Antagonist.
Di Maro, S., Di Leva, F.S., Trotta, A.M., Brancaccio, D., Portella, L., Aurilio, M., Tomassi, S., Messere, A., Sementa, D., Lastoria, S., Carotenuto, A., Novellino, E., Scala, S., Marinelli, L.(2017) J Med Chem 60: 9641-9652
- PubMed: 29125295
- DOI: https://doi.org/10.1021/acs.jmedchem.7b01062
- Primary Citation of Related Structures:
5OJT - PubMed Abstract:
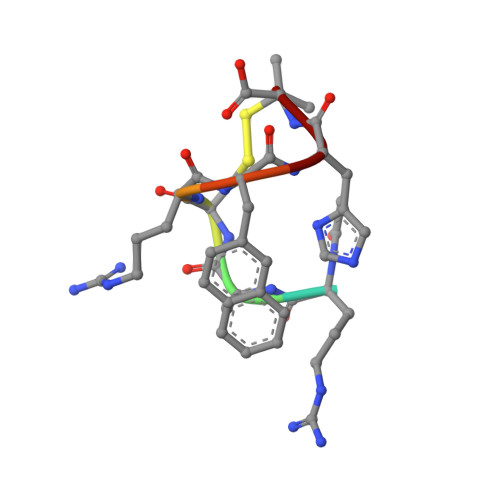
In our ongoing pursuit of CXCR4 antagonists as potential anticancer agents, we recently developed a potent, selective, and plasma stable peptide, Ac-Arg-Ala-[d-Cys-Arg-Phe-Phe-Cys]-COOH (3). Nevertheless, this compound was still not potent enough (IC 50 ≈ 53 nM) to enter preclinical studies. Thus, a lead-optimization campaign was here undertaken to further improve the binding affinity of 3 while preserving its selectivity and proteolytic stability. Specifically, extensive structure-activity relationships (SARs) investigations were carried out on both its aromatic and disulfide forming amino acids. One among the synthesized analogue, Ac-Arg-Ala-[d-Cys-Arg-Phe-His-Pen]-COOH (19), displayed subnanomolar affinity toward CXCR4, with a marked selectivity over CXCR3 and CXCR7. NMR and molecular modeling studies disclosed the molecular bases for the binding of 19 to CXCR4 and for its improved potency compared to the lead 3. Finally, biological assays on specific cancer cell lines showed that 19 can impair CXCL12-mediated cell migration and CXCR4 internalization more efficiently than the clinically approved CXCR4 antagonist plerixafor.
- DiSTABiF, University of Campania "Luigi Vanvitelli" , Caserta 81100, Italy.
Organizational Affiliation: