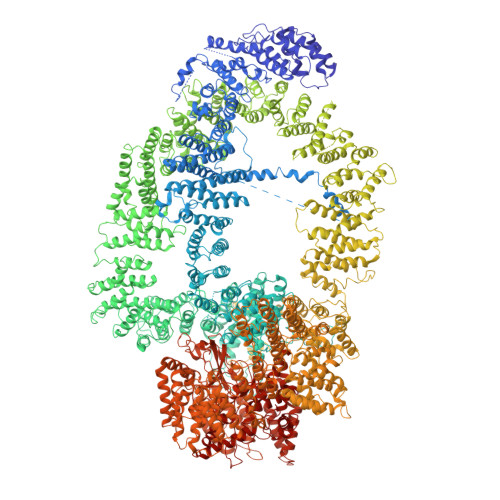
Cryo-EM structure of the SAGA and NuA4 coactivator subunit Tra1 at 3.7 angstrom resolution.
Diaz-Santin, L.M., Lukoyanova, N., Aciyan, E., Cheung, A.C.(2017) Elife 6
- PubMed: 28767037
- DOI: https://doi.org/10.7554/eLife.28384
- Primary Citation of Related Structures:
5OJS - PubMed Abstract:
Coactivator complexes SAGA and NuA4 stimulate transcription by post-translationally modifying chromatin. Both complexes contain the Tra1 subunit, a highly conserved 3744-residue protein from the Phosphoinositide 3-Kinase-related kinase (PIKK) family and a direct target for multiple sequence-specific activators. We present the Cryo-EM structure of Saccharomyces cerevsisae Tra1 to 3.7 Å resolution, revealing an extensive network of alpha-helical solenoids organized into a diamond ring conformation and is strikingly reminiscent of DNA-PKcs, suggesting a direct role for Tra1 in DNA repair. The structure was fitted into an existing SAGA EM reconstruction and reveals limited contact surfaces to Tra1, hence it does not act as a molecular scaffold within SAGA. Mutations that affect activator targeting are distributed across the Tra1 structure, but also cluster within the N-terminal Finger region, indicating the presence of an activator interaction site. The structure of Tra1 is a key milestone in deciphering the mechanism of multiple coactivator complexes.
- Department of Structural and Molecular Biology, Institute of Structural and Molecular Biology, University College London, London, United Kingdom.
Organizational Affiliation: