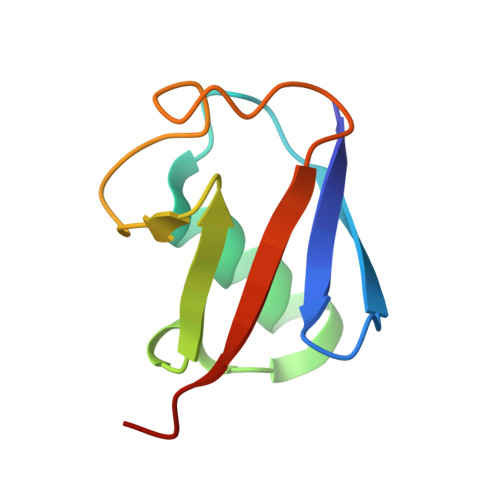
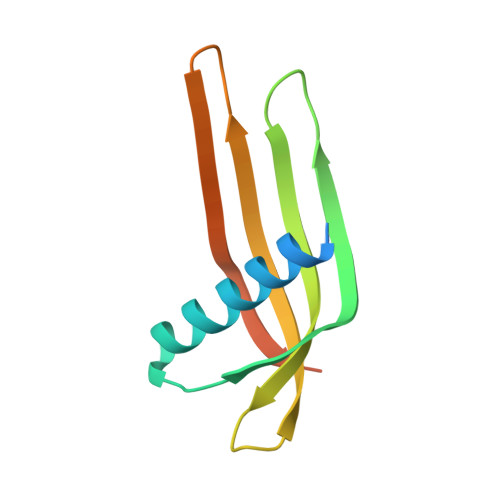
Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling.
Michel, M.A., Swatek, K.N., Hospenthal, M.K., Komander, D.(2017) Mol Cell 68: 233-246.e5
- PubMed: 28943312
- DOI: https://doi.org/10.1016/j.molcel.2017.08.020
- Primary Citation of Related Structures:
5OHL, 5OHM, 5OHV - PubMed Abstract:
Several ubiquitin chain types have remained unstudied, mainly because tools and techniques to detect these posttranslational modifications are scarce. Linkage-specific antibodies have shaped our understanding of the roles and dynamics of polyubiquitin signals but are available for only five out of eight linkage types. We here characterize K6- and K33-linkage-specific "affimer" reagents as high-affinity ubiquitin interactors. Crystal structures of affimers bound to their cognate chain types reveal mechanisms of specificity and a K11 cross-reactivity in the K33 affimer. Structure-guided improvements yield superior affinity reagents suitable for western blotting, confocal fluorescence microscopy and pull-down applications. This allowed us to identify RNF144A and RNF144B as E3 ligases that assemble K6-, K11-, and K48-linked polyubiquitin in vitro. A protocol to enrich K6-ubiquitinated proteins from cells identifies HUWE1 as a main E3 ligase for this chain type, and we show that mitofusin-2 is modified with K6-linked polyubiquitin in a HUWE1-dependent manner.
- Division of Protein and Nucleic Acid Chemistry, MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge CB2 0QH, UK.
Organizational Affiliation: