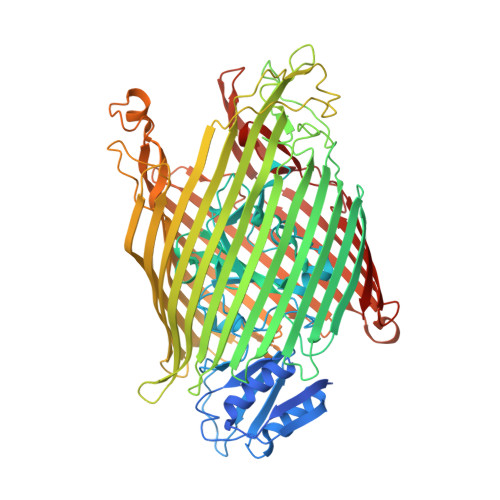
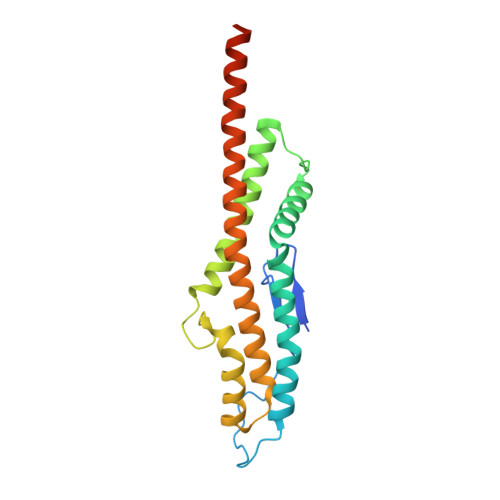
Exploitation of an iron transporter for bacterial protein antibiotic import.
White, P., Joshi, A., Rassam, P., Housden, N.G., Kaminska, R., Goult, J.D., Redfield, C., McCaughey, L.C., Walker, D., Mohammed, S., Kleanthous, C.(2017) Proc Natl Acad Sci U S A 114: 12051-12056
- PubMed: 29078392
- DOI: https://doi.org/10.1073/pnas.1713741114
- Primary Citation of Related Structures:
5ODW - PubMed Abstract:
Unlike their descendants, mitochondria and plastids, bacteria do not have dedicated protein import systems. However, paradoxically, import of protein bacteriocins, the mechanisms of which are poorly understood, underpins competition among pathogenic and commensal bacteria alike. Here, using X-ray crystallography, isothermal titration calorimetry, confocal fluorescence microscopy, and in vivo photoactivatable cross-linking of stalled translocation intermediates, we demonstrate how the iron transporter FpvAI in the opportunistic pathogen Pseudomonas aeruginosa is hijacked to translocate the bacteriocin pyocin S2 (pyoS2) across the outer membrane (OM). FpvAI is a TonB-dependent transporter (TBDT) that actively imports the small siderophore ferripyoverdine (Fe-Pvd) by coupling to the proton motive force (PMF) via the inner membrane (IM) protein TonB1. The crystal structure of the N-terminal domain of pyoS2 (pyoS2 NTD ) bound to FpvAI ( K d = 240 pM) reveals that the pyocin mimics Fe-Pvd, inducing the same conformational changes in the receptor. Mimicry leads to fluorescently labeled pyoS2 NTD being imported into FpvAI-expressing P. aeruginosa cells by a process analogous to that used by bona fide TBDT ligands. PyoS2 NTD induces unfolding by TonB1 of a force-labile portion of the plug domain that normally occludes the central channel of FpvAI. The pyocin is then dragged through this narrow channel following delivery of its own TonB1-binding epitope to the periplasm. Hence, energized nutrient transporters in bacteria also serve as rudimentary protein import systems, which, in the case of FpvAI, results in a protein antibiotic 60-fold bigger than the transporter's natural substrate being translocated across the OM.
- Department of Biochemistry, University of Oxford, Oxford OX1 3QU, United Kingdom.
Organizational Affiliation: