Nucleosome-Chd1 structure and implications for chromatin remodelling.
Farnung, L., Vos, S.M., Wigge, C., Cramer, P.(2017) Nature 550: 539-542
- PubMed: 29019976
- DOI: https://doi.org/10.1038/nature24046
- Primary Citation of Related Structures:
5O9G - PubMed Abstract:
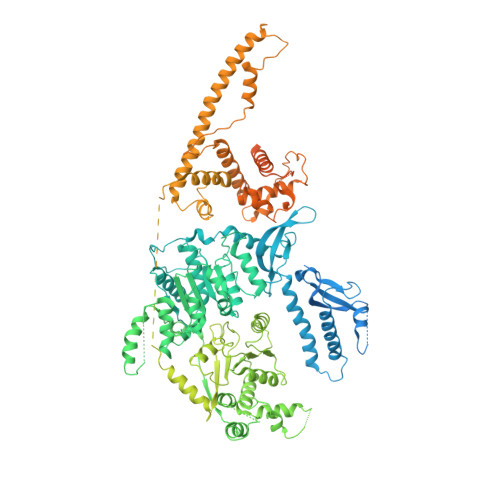
Chromatin-remodelling factors change nucleosome positioning and facilitate DNA transcription, replication, and repair. The conserved remodelling factor chromodomain-helicase-DNA binding protein 1(Chd1) can shift nucleosomes and induce regular nucleosome spacing. Chd1 is required for the passage of RNA polymerase IIthrough nucleosomes and for cellular pluripotency. Chd1 contains the DNA-binding domains SANT and SLIDE, a bilobal motor domain that hydrolyses ATP, and a regulatory double chromodomain. Here we report the cryo-electron microscopy structure of Chd1 from the yeast Saccharomyces cerevisiae bound to a nucleosome at a resolution of 4.8 Å. Chd1 detaches two turns of DNA from the histone octamer and binds between the two DNA gyres in a state poised for catalysis. The SANT and SLIDE domains contact detached DNA around superhelical location (SHL) -7 of the first DNA gyre. The ATPase motor binds the second DNA gyre at SHL +2 and is anchored to the N-terminal tail of histone H4, as seen in a recent nucleosome-Snf2 ATPase structure. Comparisons with published results reveal that the double chromodomain swings towards nucleosomal DNA at SHL +1, resulting in ATPase closure. The ATPase can then promote translocation of DNA towards the nucleosome dyad, thereby loosening the first DNA gyre and remodelling the nucleosome. Translocation may involve ratcheting of the two lobes of the ATPase, which is trapped in a pre- or post-translocation state in the absence or presence, respectively, of transition state-mimicking compounds.
- Max Planck Institute for Biophysical Chemistry, Department of Molecular Biology, Am Fassberg 11, 37077 Göttingen, Germany.
Organizational Affiliation: