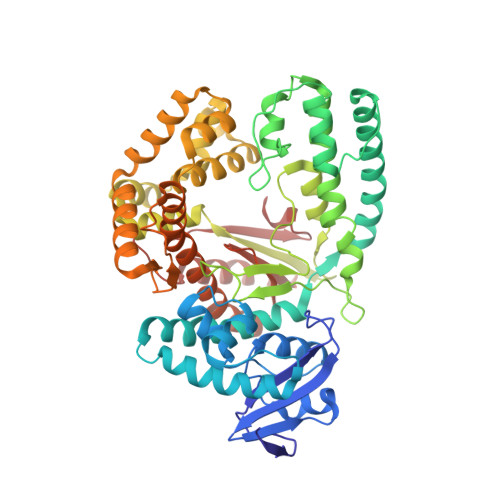
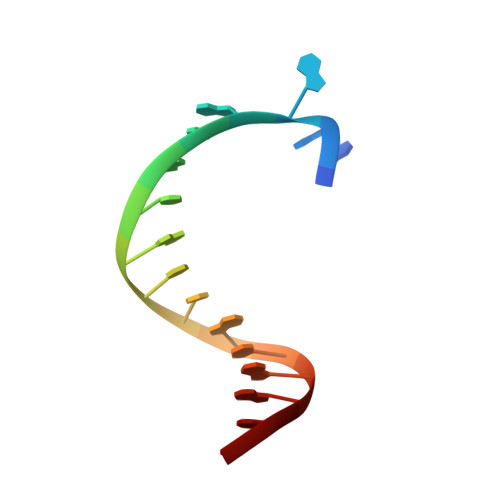
Structural basis for the selective incorporation of an artificial nucleotide opposite a DNA adduct by a DNA polymerase.
Betz, K., Nilforoushan, A., Wyss, L.A., Diederichs, K., Sturla, S.J., Marx, A.(2017) Chem Commun (Camb) 53: 12704-12707
- PubMed: 29136072
- DOI: https://doi.org/10.1039/c7cc07173f
- Primary Citation of Related Structures:
5O7T, 5OXJ - PubMed Abstract:
The possibility to sequence cytotoxic O 6 -alkylG DNA adducts would greatly benefit research. Recently we reported a benzimidazole-derived nucleotide that is selectively incorporated opposite the damaged site by a mutated DNA polymerase. Here we provide the structural basis for this reaction which may spur future developments in DNA damage sequencing.
- Department of Chemistry & Konstanz Research School Chemical Biology, University of Konstanz, Universitätsstr. 10, D-78457 Konstanz, Germany. andreas.marx@uni-konstanz.de.
Organizational Affiliation: