Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide
Fadouloglou, V.E., Kotsifaki, D., Kokkinidis, M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptidoglycan N-acetylglucosamine deacetylase | 214 | Bacillus cereus ATCC 14579 | Mutation(s): 0 Gene Names: BC_1960 EC: 3.5.1 (PDB Primary Data), 3.5.1.104 (UniProt) | 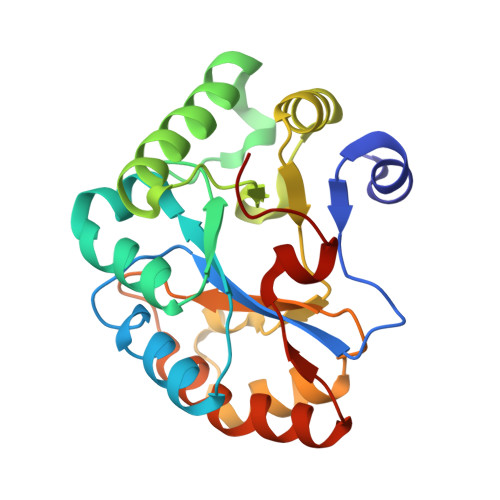 | |
UniProt | |||||
Find proteins for Q81EK9 (Bacillus cereus (strain ATCC 14579 / DSM 31 / CCUG 7414 / JCM 2152 / NBRC 15305 / NCIMB 9373 / NCTC 2599 / NRRL B-3711)) Explore Q81EK9 Go to UniProtKB: Q81EK9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q81EK9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptidoglycan N-acetylglucosamine deacetylase | 214 | Bacillus cereus ATCC 14579 | Mutation(s): 0 Gene Names: BC_1960 EC: 3.5.1 (PDB Primary Data), 3.5.1.104 (UniProt) | 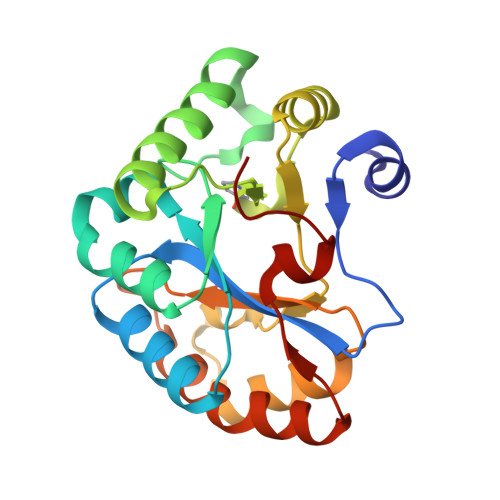 | |
UniProt | |||||
Find proteins for Q81EK9 (Bacillus cereus (strain ATCC 14579 / DSM 31 / CCUG 7414 / JCM 2152 / NBRC 15305 / NCIMB 9373 / NCTC 2599 / NRRL B-3711)) Explore Q81EK9 Go to UniProtKB: Q81EK9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q81EK9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 9 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PE3 Query on PE3 | I [auth A], UA [auth C] | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL C28 H58 O15 ILLKMACMBHTSHP-UHFFFAOYSA-N |  | ||
| 5YA Query on 5YA | CB [auth D], H [auth A], SA [auth C], W [auth B] | 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide C17 H13 N O2 PKQVYJOPUCLLCD-UHFFFAOYSA-N |  | ||
| PG0 Query on PG0 | M [auth A] | 2-(2-METHOXYETHOXY)ETHANOL C5 H12 O3 SBASXUCJHJRPEV-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | AA [auth B] BA [auth B] CA [auth B] EB [auth D] FB [auth D] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | BB [auth D] E [auth A] F [auth A] G [auth A] OA [auth C] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| DMS Query on DMS | Y [auth B], Z [auth B] | DIMETHYL SULFOXIDE C2 H6 O S IAZDPXIOMUYVGZ-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | DB [auth D], IB [auth D], TA [auth C], X [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| CL Query on CL | JB [auth D], MA [auth B], NA [auth B], U [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | AB [auth C] DA [auth B] EA [auth B] FA [auth B] GA [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| PXU Query on PXU | B, C, D | L-PEPTIDE LINKING | C5 H9 N O3 |  | PRO |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 93.042 | α = 90 |
| b = 93.042 | β = 90 |
| c = 243.868 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Union | InnovCrete |