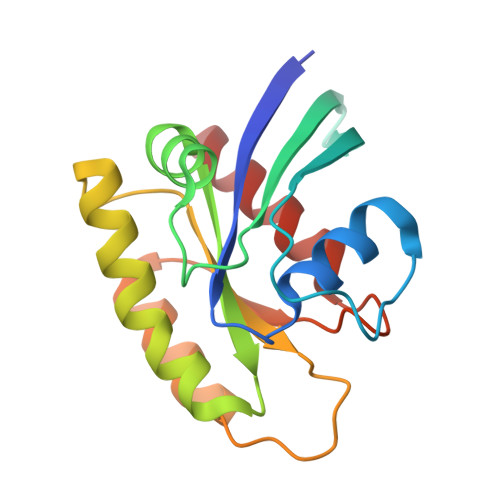
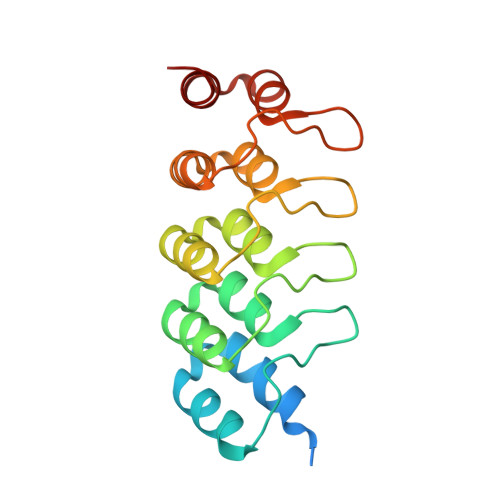
Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange.
Guillard, S., Kolasinska-Zwierz, P., Debreczeni, J., Breed, J., Zhang, J., Bery, N., Marwood, R., Tart, J., Overman, R., Stocki, P., Mistry, B., Phillips, C., Rabbitts, T., Jackson, R., Minter, R.(2017) Nat Commun 8: 16111-16111
- PubMed: 28706291
- DOI: https://doi.org/10.1038/ncomms16111
- Primary Citation of Related Structures:
5O2S, 5O2T - PubMed Abstract:
Ras mutations are the oncogenic drivers of many human cancers and yet there are still no approved Ras-targeted cancer therapies. Inhibition of Ras nucleotide exchange is a promising new approach but better understanding of this mechanism of action is needed. Here we describe an antibody mimetic, DARPin K27, which inhibits nucleotide exchange of Ras. K27 binds preferentially to the inactive Ras GDP form with a K d of 4 nM and structural studies support its selectivity for inactive Ras. Intracellular expression of K27 significantly reduces the amount of active Ras, inhibits downstream signalling, in particular the levels of phosphorylated ERK, and slows the growth in soft agar of HCT116 cells. K27 is a potent, non-covalent inhibitor of nucleotide exchange, showing consistent effects across different isoforms of Ras, including wild-type and oncogenic mutant forms.
- Antibody Discovery and Protein Engineering, MedImmune, Milstein Building, Granta Park, Cambridge CB21 6GH, UK.
Organizational Affiliation: