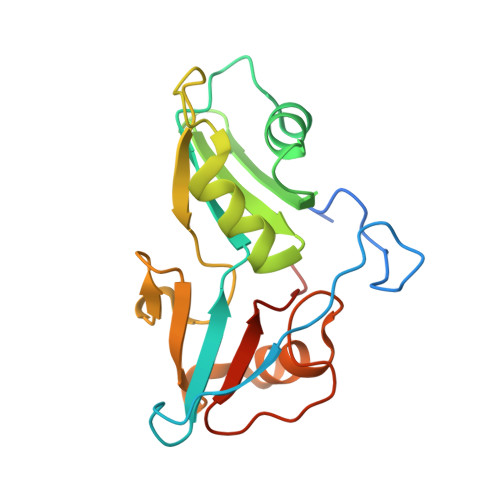
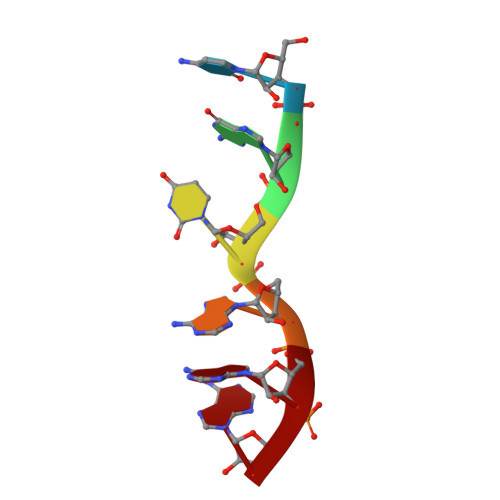
The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Franco-Echevarria, E., Gonzalez-Polo, N., Zorrilla, S., Martinez-Lumbreras, S., Santiveri, C.M., Campos-Olivas, R., Sanchez, M., Calvo, O., Gonzalez, B., Perez-Canadillas, J.M.(2017) Nucleic Acids Res 45: 10293-10305
- PubMed: 28973465
- DOI: https://doi.org/10.1093/nar/gkx685
- Primary Citation of Related Structures:
5O1T, 5O1W, 5O1X, 5O1Y, 5O1Z, 5O20 - PubMed Abstract:
Transcription termination of non-coding RNAs is regulated in yeast by a complex of three RNA binding proteins: Nrd1, Nab3 and Sen1. Nrd1 is central in this process by interacting with Rbp1 of RNA polymerase II, Trf4 of TRAMP and GUAA/G terminator sequences. We lack structural data for the last of these binding events. We determined the structures of Nrd1 RNA binding domain and its complexes with three GUAA-containing RNAs, characterized RNA binding energetics and tested rationally designed mutants in vivo. The Nrd1 structure shows an RRM domain fused with a second α/β domain that we name split domain (SD), because it is formed by two non-consecutive segments at each side of the RRM. The GUAA interacts with both domains and with a pocket of water molecules, trapped between the two stacking adenines and the SD. Comprehensive binding studies demonstrate for the first time that Nrd1 has a slight preference for GUAA over GUAG and genetic and functional studies suggest that Nrd1 RNA binding domain might play further roles in non-coding RNAs transcription termination.
- Departament of Crystallography and Structural Biology, Institute of Physical-Chemistry "Rocasolano", CSIC, C/ Serrano 119, 28006 Madrid, Spain.
Organizational Affiliation: