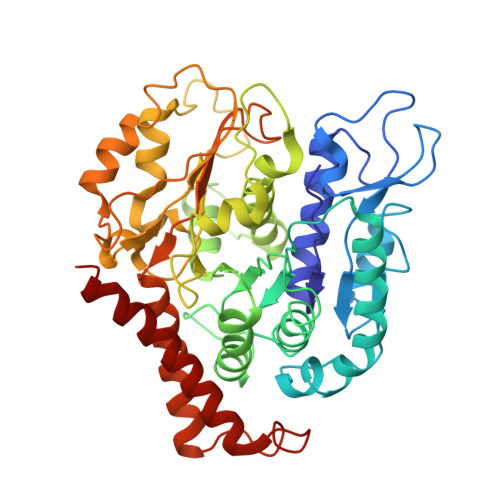
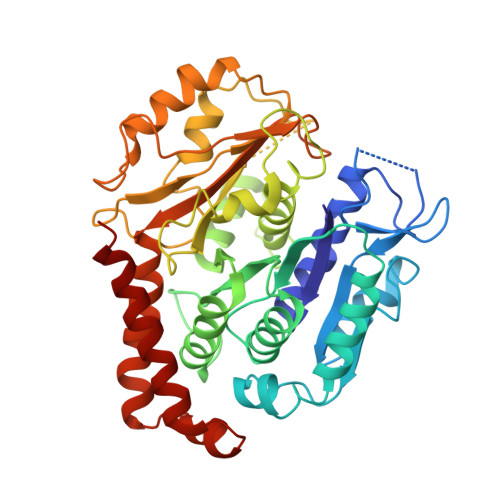
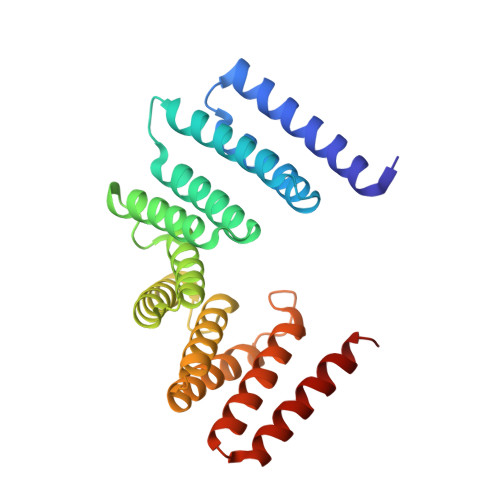
Four-stranded mini microtubules formed by Prosthecobacter BtubAB show dynamic instability.
Deng, X., Fink, G., Bharat, T.A.M., He, S., Kureisaite-Ciziene, D., Lowe, J.(2017) Proc Natl Acad Sci U S A 114: E5950-E5958
- PubMed: 28673988
- DOI: https://doi.org/10.1073/pnas.1705062114
- Primary Citation of Related Structures:
5O01, 5O09 - PubMed Abstract:
Microtubules, the dynamic, yet stiff hollow tubes built from αβ-tubulin protein heterodimers, are thought to be present only in eukaryotic cells. Here, we report a 3.6-Å helical reconstruction electron cryomicroscopy structure of four-stranded mini microtubules formed by bacterial tubulin-like Prosthecobacter dejongeii BtubAB proteins. Despite their much smaller diameter, mini microtubules share many key structural features with eukaryotic microtubules, such as an M-loop, alternating subunits, and a seam that breaks overall helical symmetry. Using in vitro total internal reflection fluorescence microscopy, we show that bacterial mini microtubules treadmill and display dynamic instability, another hallmark of eukaryotic microtubules. The third protein in the btub gene cluster, BtubC, previously known as "bacterial kinesin light chain," binds along protofilaments every 8 nm, inhibits BtubAB mini microtubule catastrophe, and increases rescue. Our work reveals that some bacteria contain regulated and dynamic cytomotive microtubule systems that were once thought to be only useful in much larger and sophisticated eukaryotic cells.
- Structural Studies Division, Medical Research Council Laboratory of Molecular Biology, Cambridge CB2 0QH, United Kingdom.
Organizational Affiliation: