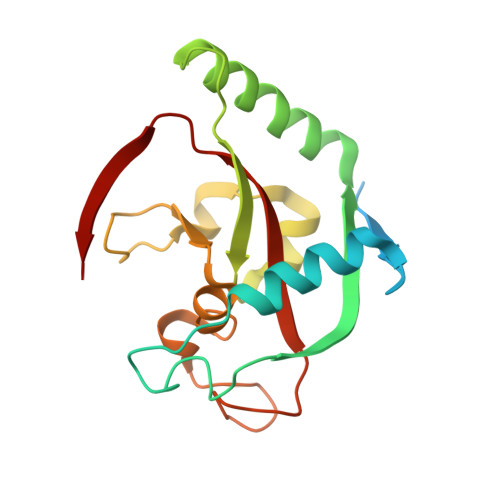
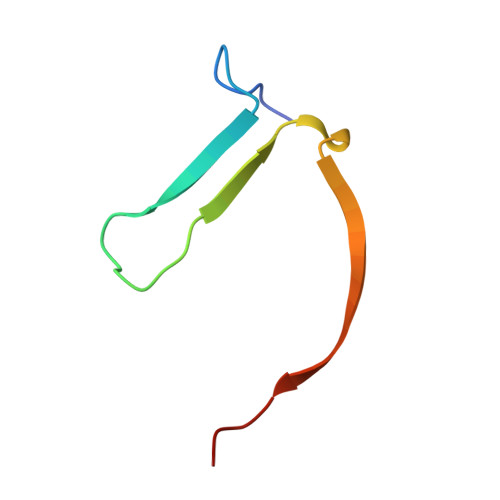
2-Phenylquinazolinones as dual-activity tankyrase-kinase inhibitors.
Nkizinkiko, Y., Desantis, J., Koivunen, J., Haikarainen, T., Murthy, S., Sancineto, L., Massari, S., Ianni, F., Obaji, E., Loza, M.I., Pihlajaniemi, T., Brea, J., Tabarrini, O., Lehtio, L.(2018) Sci Rep 8: 1680-1680
- PubMed: 29374194
- DOI: https://doi.org/10.1038/s41598-018-19872-3
- Primary Citation of Related Structures:
5NSX, 5NT0, 5NT4, 5NUT, 5NVC, 5NVE, 5NVF, 5NVH, 5NWB, 5NWC, 5NWD, 5NWG, 5NXE, 5OWS, 5OWT - PubMed Abstract:
Tankyrases (TNKSs) are enzymes specialized in catalyzing poly-ADP-ribosylation of target proteins. Several studies have validated TNKSs as anti-cancer drug targets due to their regulatory role in Wnt/β-catenin pathway. Recently a lot of effort has been put into developing more potent and selective TNKS inhibitors and optimizing them towards anti-cancer agents. We noticed that some 2-phenylquinazolinones (2-PQs) reported as CDK9 inhibitors were similar to previously published TNKS inhibitors. In this study, we profiled this series of 2-PQs against TNKS and selected kinases that are involved in the Wnt/β-catenin pathway. We found that they were much more potent TNKS inhibitors than they were CDK9/kinase inhibitors. We evaluated the compound selectivity to tankyrases over the ARTD enzyme family and solved co-crystal structures of the compounds with TNKS2. Comparative structure-based studies of the catalytic domain of TNKS2 with selected CDK9 inhibitors and docking studies of the inhibitors with two kinases (CDK9 and Akt) revealed important structural features, which could explain the selectivity of the compounds towards either tankyrases or kinases. We also discovered a compound, which was able to inhibit tankyrases, CDK9 and Akt kinases with equal µM potency.
- Faculty of Biochemistry and Molecular Medicine & Biocenter Oulu, University of Oulu, Oulu, Finland.
Organizational Affiliation: