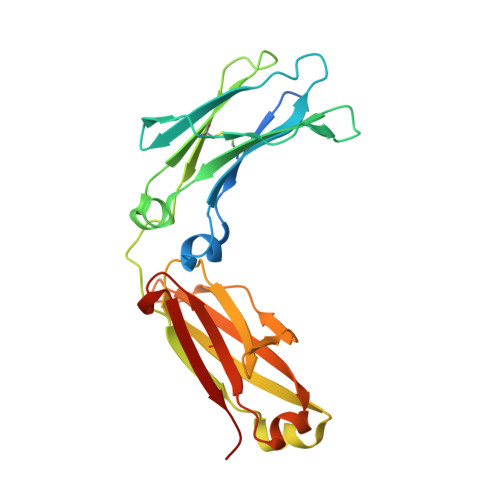
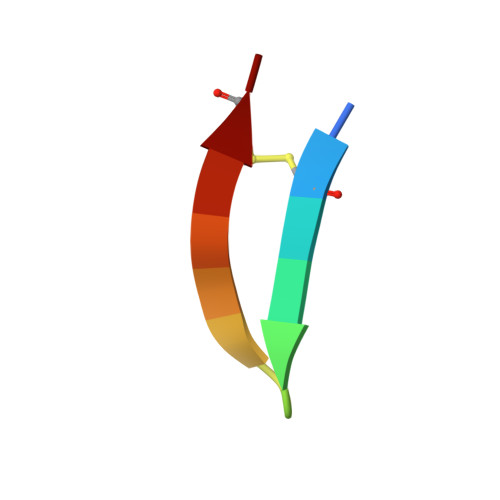
A new approach for generating bispecific antibodies based on a common light chain format and the stable architecture of human immunoglobulin G1.
De Nardis, C., Hendriks, L.J.A., Poirier, E., Arvinte, T., Gros, P., Bakker, A.B.H., de Kruif, J.(2017) J Biological Chem 292: 14706-14717
- PubMed: 28655766
- DOI: https://doi.org/10.1074/jbc.M117.793497
- Primary Citation of Related Structures:
5NSC, 5NSG - PubMed Abstract:
Bispecific antibodies combine two different antigen-binding sites in a single molecule, enabling more specific targeting, novel mechanisms of action, and higher clinical efficacies. Although they have the potential to outperform conventional monoclonal antibodies, many bispecific antibodies have issues regarding production, stability, and pharmacokinetic properties. Here, we describe a new approach for generating bispecific antibodies using a common light chain format and exploiting the stable architecture of human immunoglobulin G 1 We used iterative experimental validation and computational modeling to identify multiple Fc variant pairs that drive efficient heterodimerization of the antibody heavy chains. Accelerated stability studies enabled selection of one Fc variant pair dubbed "DEKK" consisting of substitutions L351D and L368E in one heavy chain combined with L351K and T366K in the other. Solving the crystal structure of the DEKK Fc region at a resolution of 2.3 Å enabled detailed analysis of the interactions inducing CH3 interface heterodimerization. Local shifts in the IgG backbone accommodate the introduction of lysine side chains that form stabilizing salt-bridge interactions with substituted and native residues in the opposite chain. Overall, the CH3 domain adapted to these shifts at the interface, yielding a stable Fc conformation very similar to that in wild-type IgG. Using the DEKK format, we generated the bispecific antibody MCLA-128, targeting human EGF receptors 2 and 3. MCLA-128 could be readily produced and purified at industrial scale with a standard mammalian cell culture platform and a routine purification protocol. Long-term accelerated stability assays confirmed that MCLA-128 is highly stable and has excellent biophysical characteristics.
- From the Crystal and Structural Chemistry Group, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Science, Utrecht University, 3584 CH Utrecht, The Netherlands.
Organizational Affiliation: