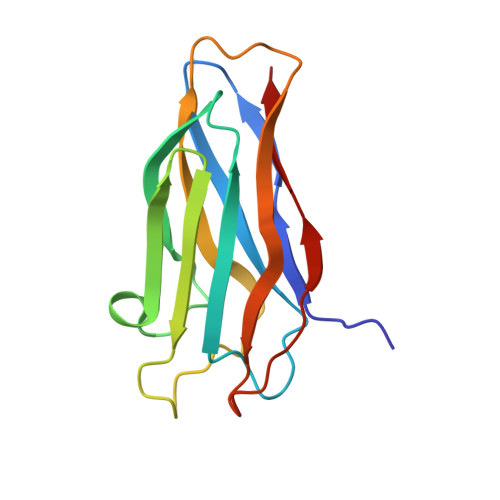
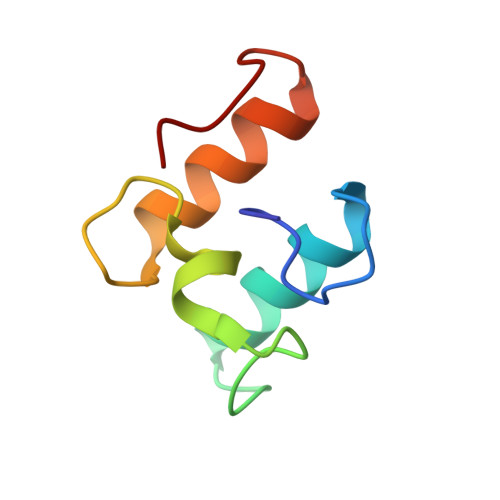
Structure-function analyses generate novel specificities to assemble the components of multienzyme bacterial cellulosome complexes.
Bule, P., Cameron, K., Prates, J.A.M., Ferreira, L.M.A., Smith, S.P., Gilbert, H.J., Bayer, E.A., Najmudin, S., Fontes, C.M.G.A., Alves, V.D.(2018) J Biological Chem 293: 4201-4212
- PubMed: 29367338
- DOI: https://doi.org/10.1074/jbc.RA117.001241
- Primary Citation of Related Structures:
5NRK, 5NRM - PubMed Abstract:
The cellulosome is a remarkably intricate multienzyme nanomachine produced by anaerobic bacteria to degrade plant cell wall polysaccharides. Cellulosome assembly is mediated through binding of enzyme-borne dockerin modules to cohesin modules of the primary scaffoldin subunit. The anaerobic bacterium Acetivibrio cellulolyticus produces a highly intricate cellulosome comprising an adaptor scaffoldin, ScaB, whose cohesins interact with the dockerin of the primary scaffoldin (ScaA) that integrates the cellulosomal enzymes. The ScaB dockerin selectively binds to cohesin modules in ScaC that anchors the cellulosome onto the cell surface. Correct cellulosome assembly requires distinct specificities displayed by structurally related type-I cohesin-dockerin pairs that mediate ScaC-ScaB and ScaA-enzyme assemblies. To explore the mechanism by which these two critical protein interactions display their required specificities, we determined the crystal structure of the dockerin of a cellulosomal enzyme in complex with a ScaA cohesin. The data revealed that the enzyme-borne dockerin binds to the ScaA cohesin in two orientations, indicating two identical cohesin-binding sites. Combined mutagenesis experiments served to identify amino acid residues that modulate type-I cohesin-dockerin specificity in A. cellulolyticus Rational design was used to test the hypothesis that the ligand-binding surfaces of ScaA- and ScaB-associated dockerins mediate cohesin recognition, independent of the structural scaffold. Novel specificities could thus be engineered into one, but not both, of the ligand-binding sites of ScaB, whereas attempts at manipulating the specificity of the enzyme-associated dockerin were unsuccessful. These data indicate that dockerin specificity requires critical interplay between the ligand-binding surface and the structural scaffold of these modules.
- From the CIISA-Faculdade de Medicina Veterinária, ULisboa, Pólo Universitário do Alto da Ajuda, Avenida da Universidade Técnica, 1300-477 Lisboa, Portugal.
Organizational Affiliation: