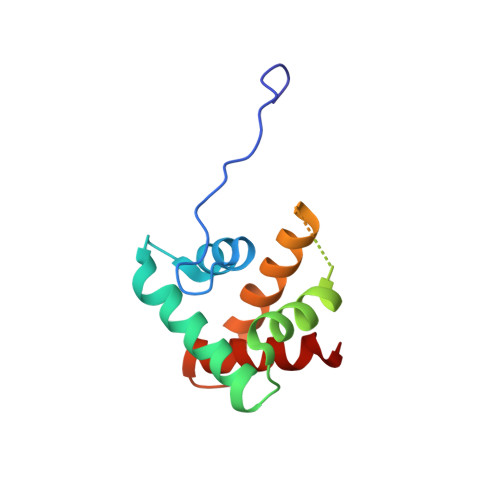
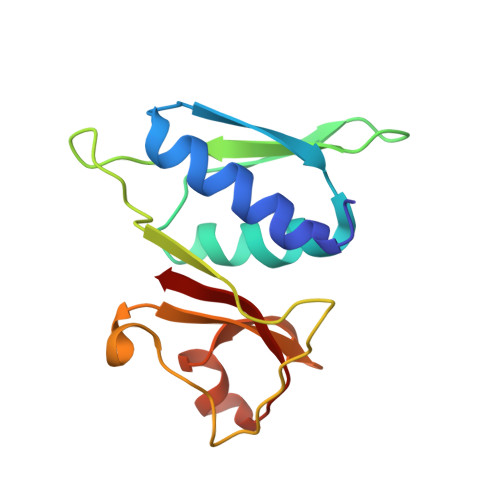
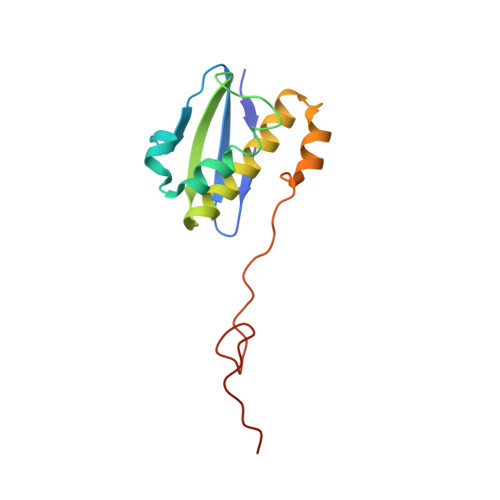
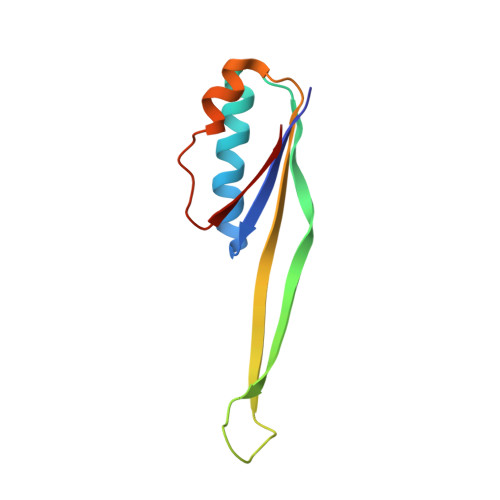
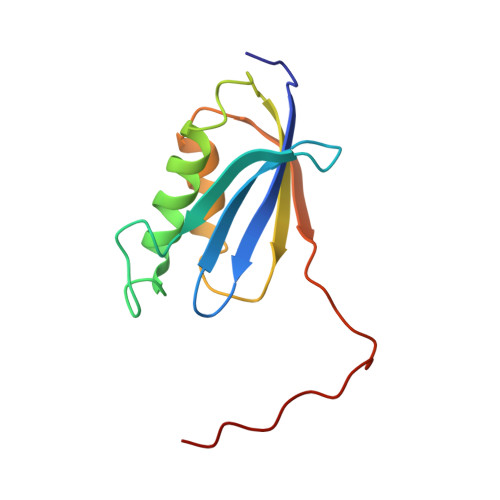
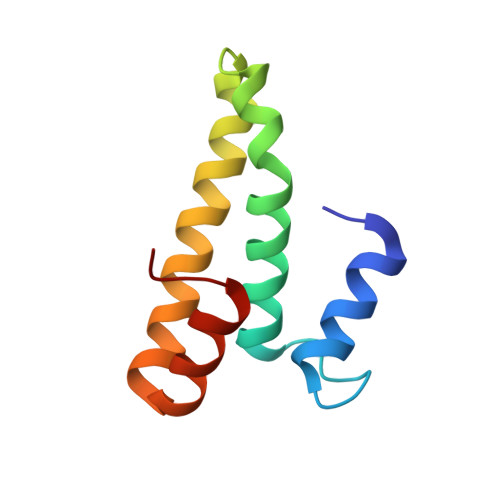
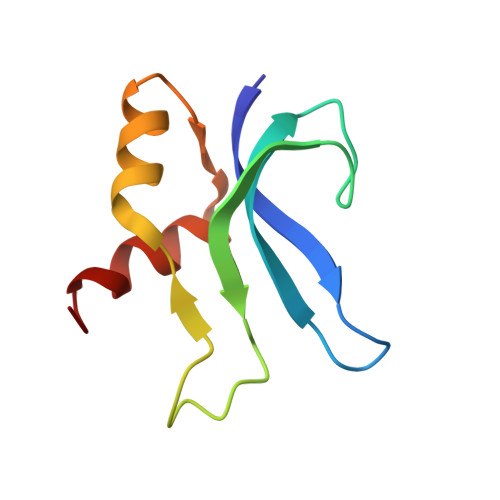
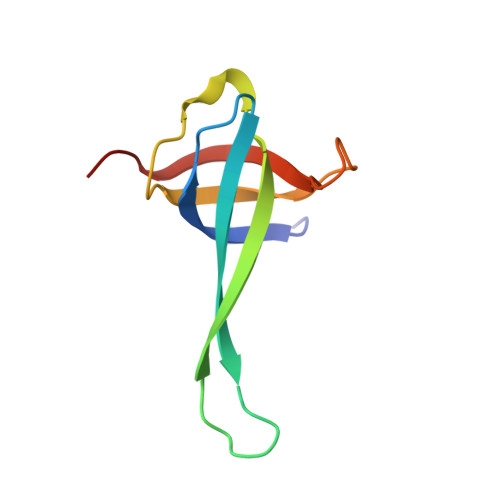
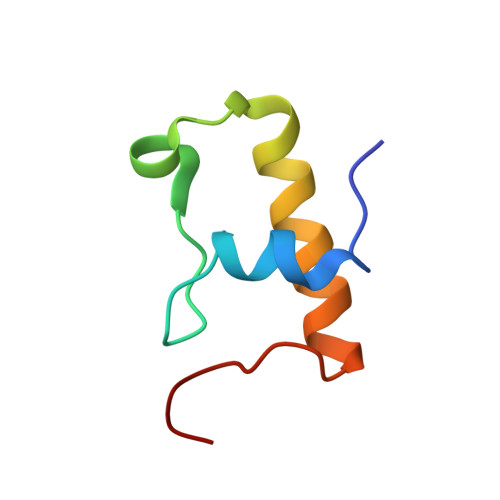
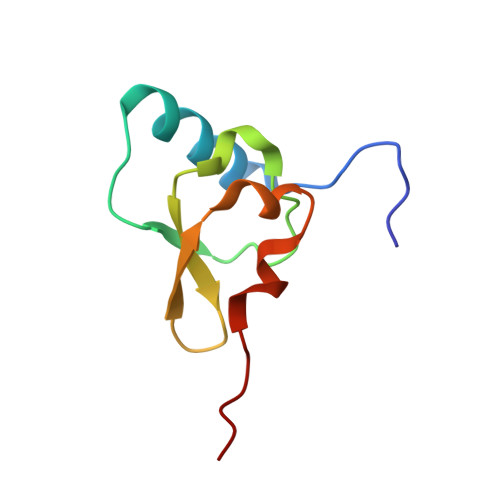
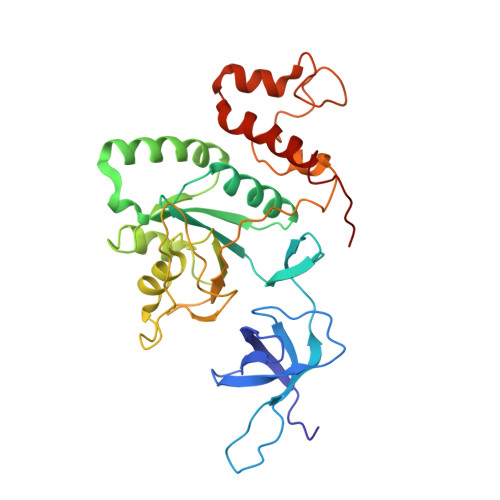
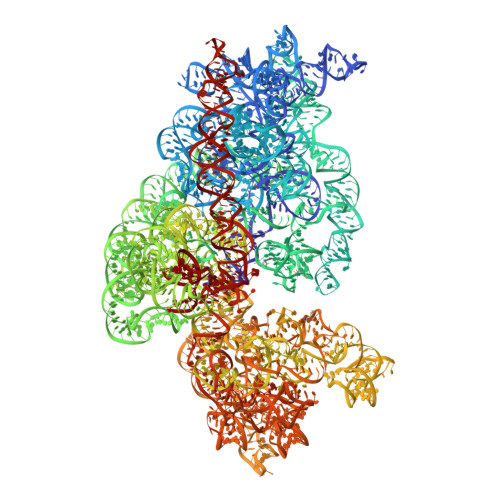
RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Lopez-Alonso, J.P., Kaminishi, T., Kikuchi, T., Hirata, Y., Iturrioz, I., Dhimole, N., Schedlbauer, A., Hase, Y., Goto, S., Kurita, D., Muto, A., Zhou, S., Naoe, C., Mills, D.J., Gil-Carton, D., Takemoto, C., Himeno, H., Fucini, P., Connell, S.R.(2017) Nucleic Acids Res 45: 6945-6959
- PubMed: 28482099
- DOI: https://doi.org/10.1093/nar/gkx324
- Primary Citation of Related Structures:
5NO2, 5NO3, 5NO4 - PubMed Abstract:
During 30S ribosomal subunit biogenesis, assembly factors are believed to prevent accumulation of misfolded intermediate states of low free energy that slowly convert into mature 30S subunits, namely, kinetically trapped particles. Among the assembly factors, the circularly permuted GTPase, RsgA, plays a crucial role in the maturation of the 30S decoding center. Here, directed hydroxyl radical probing and single particle cryo-EM are employed to elucidate RsgA΄s mechanism of action. Our results show that RsgA destabilizes the 30S structure, including late binding r-proteins, providing a structural basis for avoiding kinetically trapped assembly intermediates. Moreover, RsgA exploits its distinct GTPase pocket and specific interactions with the 30S to coordinate GTPase activation with the maturation state of the 30S subunit. This coordination validates the architecture of the decoding center and facilitates the timely release of RsgA to control the progression of 30S biogenesis.
- Molecular Recognition and Host-Pathogen Interactions, CIC bioGUNE, Bizkaia Technology Park, 48160 Derio, Spain.
Organizational Affiliation: