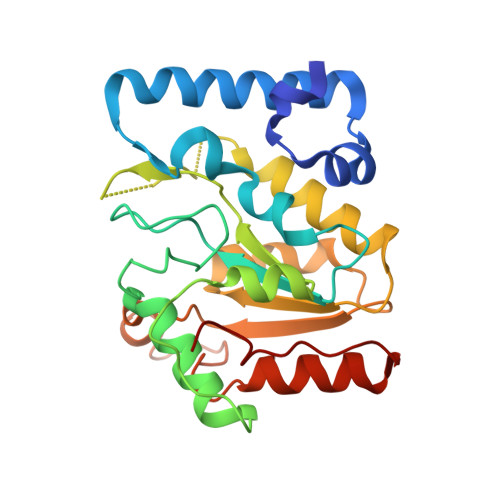
A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Earl, C., Bagneris, C., Zeman, K., Cole, A., Barrett, T., Savva, R.(2018) Nucleic Acids Res 46: 4286-4300
- PubMed: 29596604
- DOI: https://doi.org/10.1093/nar/gky217
- Primary Citation of Related Structures:
5NN7, 5NNH, 5NNU - PubMed Abstract:
Efficient γ-herpesvirus lytic phase replication requires a virally encoded UNG-type uracil-DNA glycosylase as a structural element of the viral replisome. Uniquely, γ-herpesvirus UNGs carry a seven or eight residue insertion of variable sequence in the otherwise highly conserved minor-groove DNA binding loop. In Epstein-Barr Virus [HHV-4] UNG, this motif forms a disc-shaped loop structure of unclear significance. To ascertain the biological role of the loop insertion, we determined the crystal structure of Kaposi's sarcoma-associated herpesvirus [HHV-8] UNG (kUNG) in its product complex with a uracil-containing dsDNA, as well as two structures of kUNG in its apo state. We find the disc-like conformation is conserved, but only when the kUNG DNA-binding cleft is occupied. Surprisingly, kUNG uses this structure to flip the orphaned partner base of the substrate deoxyuridine out of the DNA duplex while retaining canonical UNG deoxyuridine-flipping and catalysis. The orphan base is stably posed in the DNA major groove which, due to DNA backbone manipulation by kUNG, is more open than in other UNG-dsDNA structures. Mutagenesis suggests a model in which the kUNG loop is pinned outside the DNA-binding cleft until DNA docking promotes rigid structuring of the loop and duplex nucleotide flipping, a novel observation for UNGs.
- Department of Biological Sciences, Institute of Structural and Molecular Biology, Birkbeck College, University of London, Malet Street, London WC1E 7HX, UK.
Organizational Affiliation: