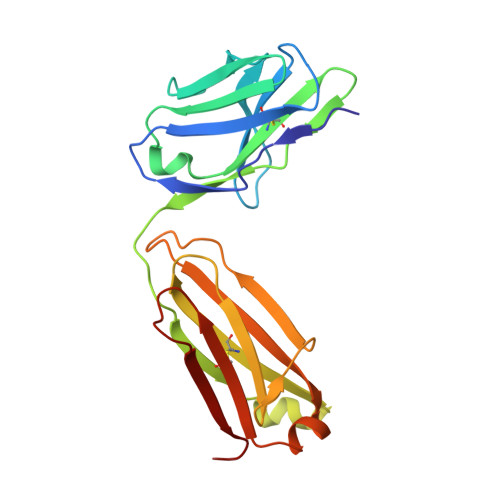
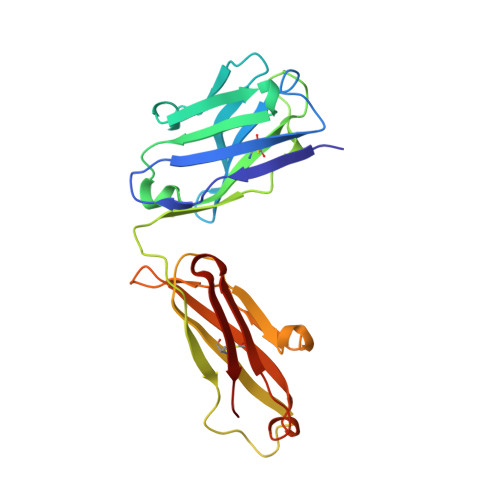
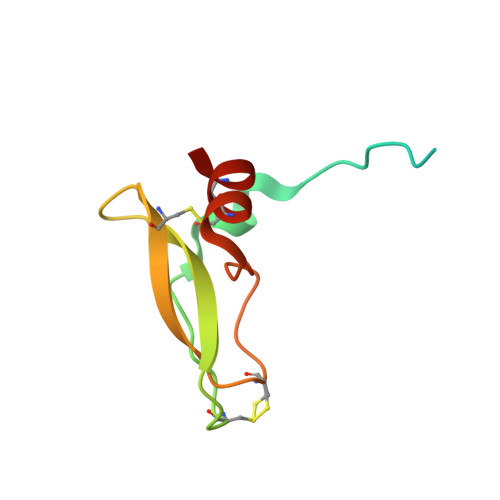
Factor Xa and VIIa inhibition by tissue factor pathway inhibitor is prevented by a monoclonal antibody to its Kunitz-1 domain.
Augustsson, C., Svensson, A., Kjaer, B., Chao, T.Y., Wenjuan, X., Krogh, B.O., Breinholt, J., Clausen, J.T., Hilden, I., Petersen, H.H., Petersen, L.C.(2018) J Thromb Haemost 16: 893-904
- PubMed: 29532595
- DOI: https://doi.org/10.1111/jth.14000
- Primary Citation of Related Structures:
5NMV - PubMed Abstract:
Essentials Activated FVII (FVIIa) and FX (FXa) are inhibited by tissue factor pathway inhibitor (TFPI). A monoclonal antibody, mAb2F22, was raised against the N-terminal fragment of TFPI (1-79). mAb2F22 bound exclusively to the K1 domain of TFPI (K D ∼1 nm) and not to the K2 domain. mAb2F22 interfered with inhibition of both FVIIa and FXa activities and restored clot formation. Background Initiation of coagulation is induced by binding of activated factor VII (FVIIa) to tissue factor (TF) and activation of factor X (FX) in a process regulated by tissue factor pathway inhibitor (TFPI). TFPI contains three Kunitz-type protease inhibitor domains (K1-K3), of which K1 and K2 block the active sites of FVIIa and FXa, respectively. Objective To produce a monoclonal antibody (mAb) directed towards K1, to characterize the binding epitope, and to study its effect on TFPI inhibition. Methods A monoclonal antibody, mAb2F22, was raised against the N-terminal TFPI(1-79) fragment. Binding data were obtained by surface plasmon resonance analysis. The Fab-fragment of mAb2F22, Fab2F22, was expressed and the structure of its complex with TFPI(1-79) determined by X-ray crystallography. Effects of mAb2F22 on TFPI inhibition were measured in buffer- and plasma-based systems. Results mAb2F22 bound exclusively to K1 of TFPI (K D ~1 nm) and not to K2. The crystal structure of Fab2F22/TFPI (1-79) mapped an epitope on K1 including seven residues upstream of the domain. TFPI inhibition of TF/FVIIa amidolytic activity was neutralized by mAb2F22, although the binding epitope on K1 did not include the P1 residue. Binding of mAb2F22 to K1 blocked TFPI inhibition of the FXa amidolytic activity and normalized hemostasis in hemophilia human A-like plasma and whole blood. Conclusion mAb2F22 blocked TFPI inhibition of both FVIIa and FXa activities and mapped a FXa exosite for binding to K1. It reversed TFPI feedback inhibition of TF/FVIIa-induced coagulation and restored clot formation in FVIII-neutralized human plasma and blood.
- Global Research, Novo Nordisk A/S, Måløv, Denmark.
Organizational Affiliation: