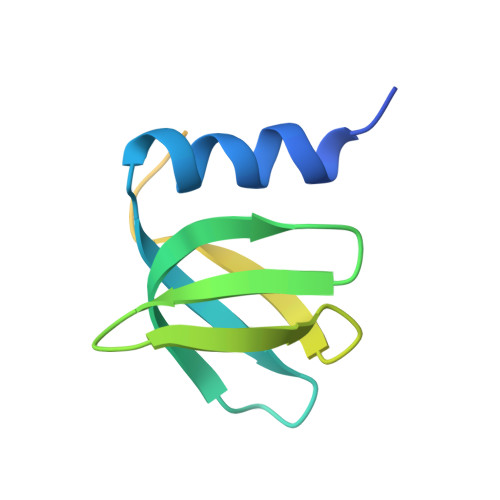
Intermolecular base stacking mediates RNA-RNA interaction in a crystal structure of the RNA chaperone Hfq.
Schulz, E.C., Seiler, M., Zuliani, C., Voigt, F., Rybin, V., Pogenberg, V., Mucke, N., Wilmanns, M., Gibson, T.J., Barabas, O.(2017) Sci Rep 7: 9903-9903
- PubMed: 28852099
- DOI: https://doi.org/10.1038/s41598-017-10085-8
- Primary Citation of Related Structures:
5NEW - PubMed Abstract:
The RNA-chaperone Hfq catalyses the annealing of bacterial small RNAs (sRNAs) with target mRNAs to regulate gene expression in response to environmental stimuli. Hfq acts on a diverse set of sRNA-mRNA pairs using a variety of different molecular mechanisms. Here, we present an unusual crystal structure showing two Hfq-RNA complexes interacting via their bound RNA molecules. The structure contains two Hfq 6 :A 18 RNA assemblies positioned face-to-face, with the RNA molecules turned towards each other and connected via interdigitating base stacking interactions at the center. Biochemical data further confirm the observed interaction, and indicate that RNA-mediated contacts occur between Hfq-RNA complexes with various (ARN) X motif containing RNA sequences in vitro, including the stress response regulator OxyS and its target, fhlA. A systematic computational survey also shows that phylogenetically conserved (ARN) X motifs are present in a subset of sRNAs, some of which share similar modular architectures. We hypothesise that Hfq can co-opt RNA-RNA base stacking, an unanticipated structural trick, to promote the interaction of (ARN) X motif containing sRNAs with target mRNAs on a "speed-dating" fashion, thereby supporting their regulatory function.
- Structural and Computational Biology Unit, European Molecular Biology Laboratory, 69117, Heidelberg, Germany.
Organizational Affiliation: