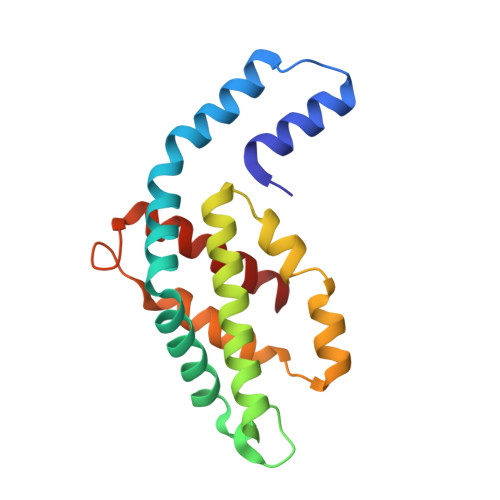
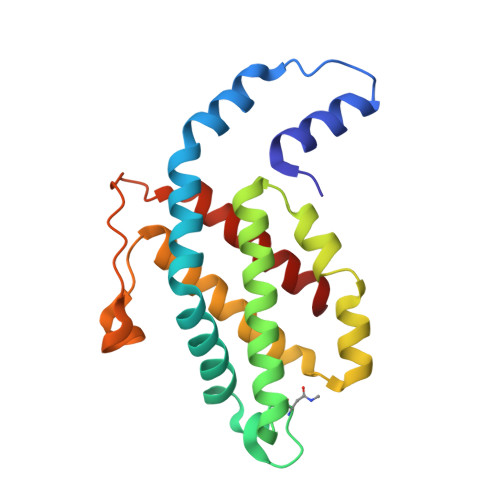
An improved crystal structure of C-phycoerythrin from the marine cyanobacterium Phormidium sp. A09DM.
Sonani, R.R., Roszak, A.W., Ortmann de Percin Northumberland, C., Madamwar, D., Cogdell, R.J.(2018) Photosynth Res 135: 65-78
- PubMed: 28918447
- DOI: https://doi.org/10.1007/s11120-017-0443-2
- Primary Citation of Related Structures:
5NB3, 5NB4 - PubMed Abstract:
C-Phycoerythrin (PE) from Phormidium sp. A09DM has been crystallized using different conditions and its structure determined to atomic resolution (1.14 Å). In order for the pigment present, phycoerythrobilin (PEB), to function as an efficient light-harvesting molecule it must be held rigidly (Kupka and Scheer in Biochim Biophys Acta 1777:94-103, 2008) and, moreover, the different PEB molecules in PE must be arranged, relative to each other, so as to promote efficient energy transfer between them. This improved structure has allowed us to define in great detail the structure of the PEBs and their binding sites. These precise structural details will facilitate theoretical calculations of each PEB's spectroscopic properties. It was possible, however, to suggest a model for which chromophores contribute to the different regions of absorption spectrum and propose a tentative scheme for energy transfer. We show that some subtle differences in one of these PEB binding sites in two of the 12 subunits are caused by crystal contacts between neighboring hexamers in the crystal lattice. This explains some of the differences seen in previous lower resolution structures determined at two different pH values (Kumar et al. in Photosyn Res 129:17-28, 2016).
- Post-Graduate Department of Biosciences, UGC-Centre of Advanced Study, Sardar Patel University, Vadtal Road, Satellite Campus, Bakrol, Anand, Gujarat, 388315, India.
Organizational Affiliation: