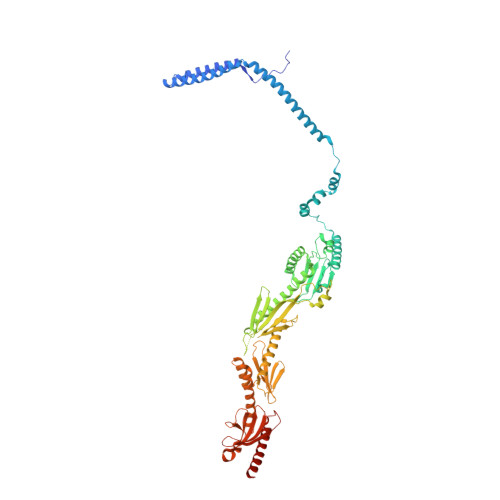
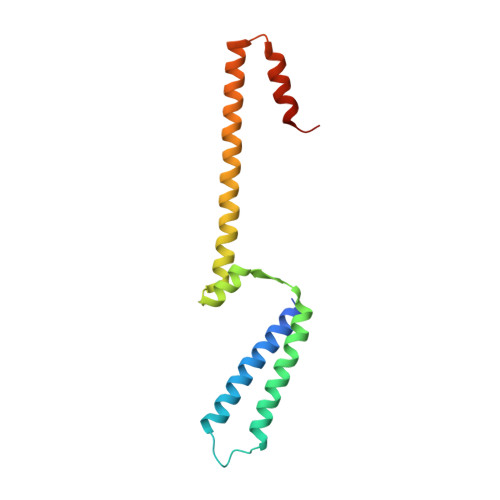
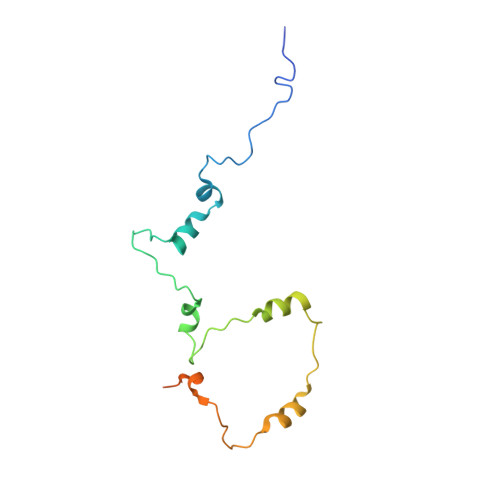
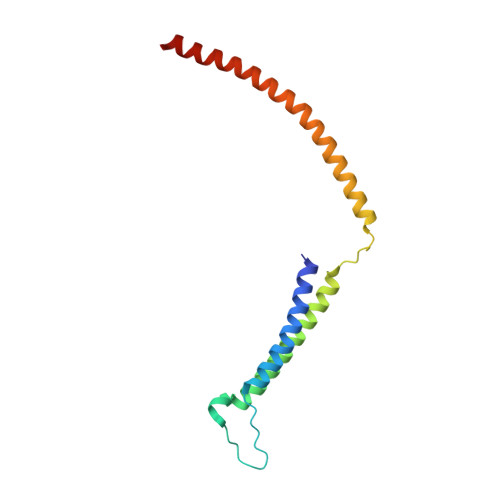
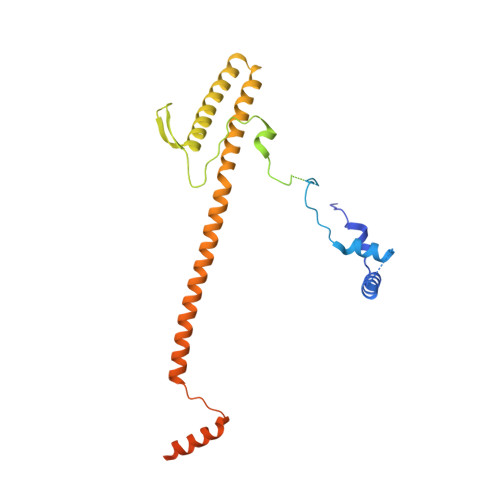
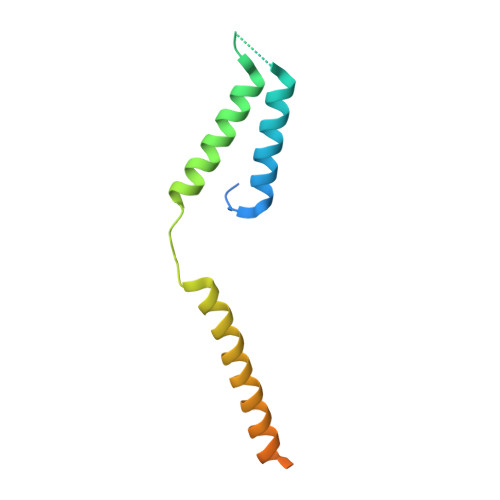
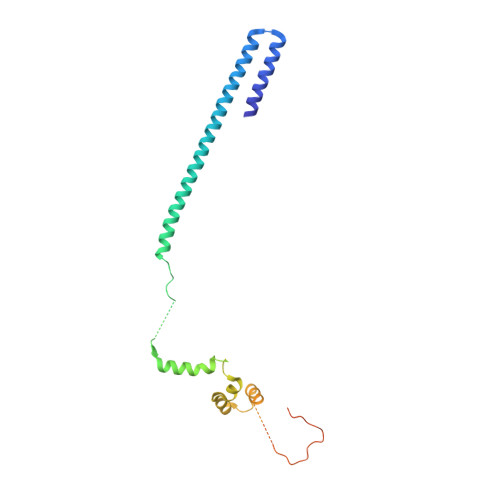
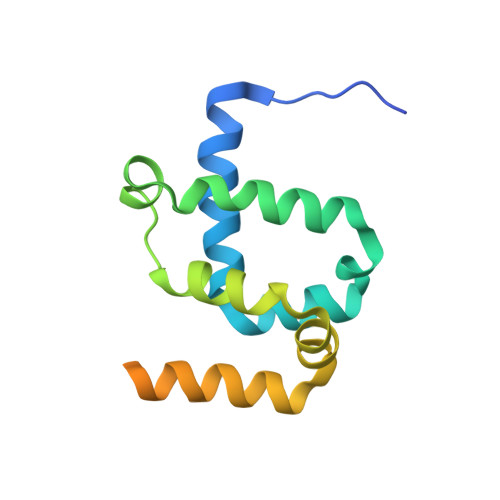
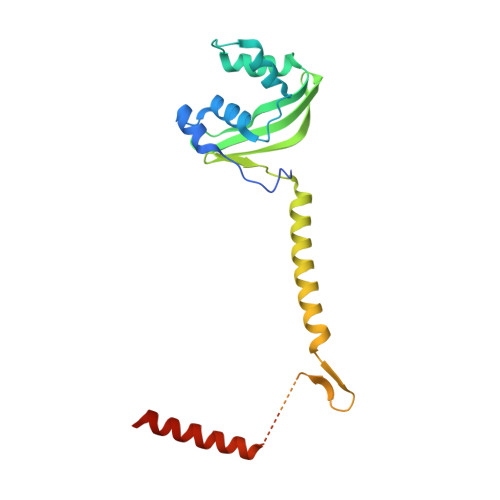
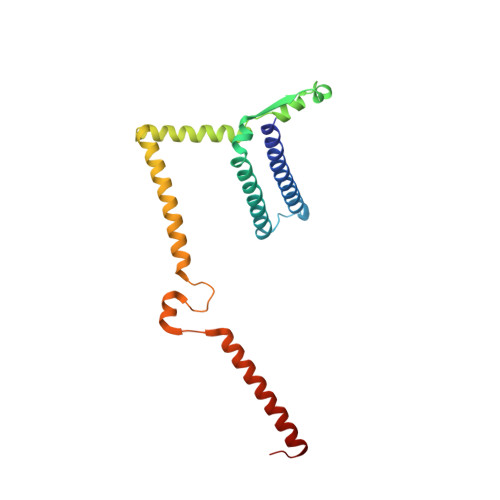
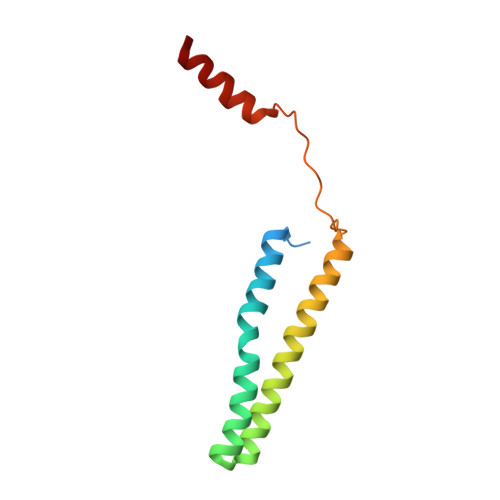
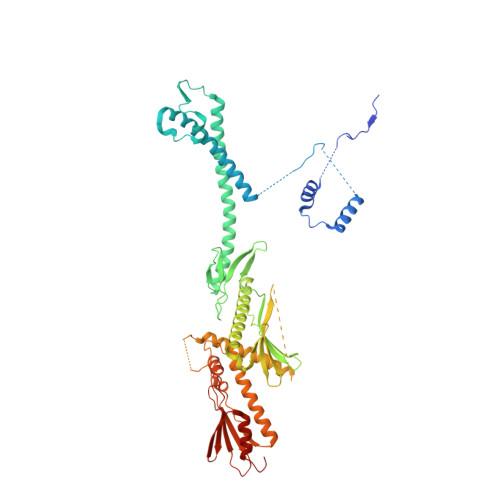
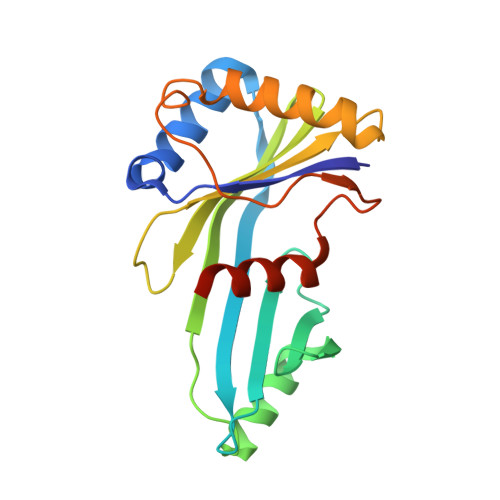
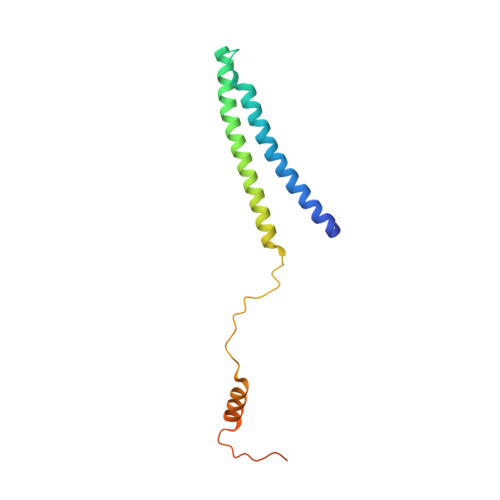
Core Mediator structure at 3.4 Angstrom extends model of transcription initiation complex.
Nozawa, K., Schneider, T.R., Cramer, P.(2017) Nature 545: 248-251
- PubMed: 28467824
- DOI: https://doi.org/10.1038/nature22328
- Primary Citation of Related Structures:
5N9J - PubMed Abstract:
Mediator is a multiprotein co-activator that binds the transcription pre-initiation complex (PIC) and regulates RNA polymerase (Pol) II. The Mediator head and middle modules form the essential core Mediator (cMed), whereas the tail and kinase modules play regulatory roles. The architecture of Mediator and its position on the PIC are known, but atomic details are limited to Mediator subcomplexes. Here we report the crystal structure of the 15-subunit cMed from Schizosaccharomyces pombe at 3.4 Å resolution. The structure shows an unaltered head module, and reveals the intricate middle module, which we show is globally required for transcription. Sites of known Mediator mutations cluster at the interface between the head and middle modules, and in terminal regions of the head subunits Med6 (ref. 16) and Med17 (ref. 17) that tether the middle module. The structure led to a model for Saccharomyces cerevisiae cMed that could be combined with the 3.6 Å cryo-electron microscopy structure of the core PIC (cPIC). The resulting atomic model of the cPIC-cMed complex informs on interactions of the submodules forming the middle module, called beam, knob, plank, connector, and hook. The hook is flexibly linked to Mediator by a conserved hinge and contacts the transcription initiation factor IIH (TFIIH) kinase that phosphorylates the carboxy (C)-terminal domain (CTD) of Pol II and was recently positioned on the PIC. The hook also contains residues that crosslink to the CTD and reside in a previously described cradle. These results provide a framework for understanding Mediator function, including its role in stimulating CTD phosphorylation by TFIIH.
- Max Planck Institute for Biophysical Chemistry, Department of Molecular Biology, Am Fassberg 11, 37077 Göttingen, Germany.
Organizational Affiliation: