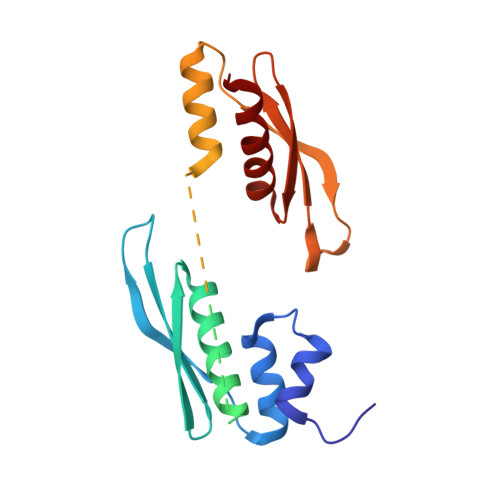
Structural basis of siRNA recognition by TRBP double-stranded RNA binding domains.
Masliah, G., Maris, C., Konig, S.L., Yulikov, M., Aeschimann, F., Malinowska, A.L., Mabille, J., Weiler, J., Holla, A., Hunziker, J., Meisner-Kober, N., Schuler, B., Jeschke, G., Allain, F.H.(2018) EMBO J 37
- PubMed: 29449323
- DOI: https://doi.org/10.15252/embj.201797089
- Primary Citation of Related Structures:
5N8L, 5N8M - PubMed Abstract:
The accurate cleavage of pre-micro(mi)RNAs by Dicer and mi/siRNA guide strand selection are important steps in forming the RNA-induced silencing complex (RISC). The role of Dicer binding partner TRBP in these processes remains poorly understood. Here, we solved the solution structure of the two N-terminal dsRNA binding domains (dsRBDs) of TRBP in complex with a functionally asymmetric siRNA using NMR, EPR, and single-molecule spectroscopy. We find that siRNA recognition by the dsRBDs is not sequence-specific but rather depends on the RNA shape. The two dsRBDs can swap their binding sites, giving rise to two equally populated, pseudo-symmetrical complexes, showing that TRBP is not a primary sensor of siRNA asymmetry. Using our structure to model a Dicer-TRBP-siRNA ternary complex, we show that TRBP's dsRBDs and Dicer's RNase III domains bind a canonical 19 base pair siRNA on opposite sides, supporting a mechanism whereby TRBP influences Dicer-mediated cleavage accuracy by binding the dsRNA region of the pre-miRNA during Dicer cleavage.
- Institute of Molecular Biology and Biophysics, ETH Zürich, Zürich, Switzerland.
Organizational Affiliation: