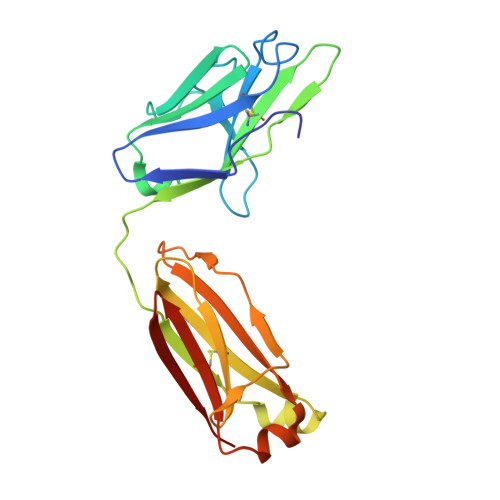
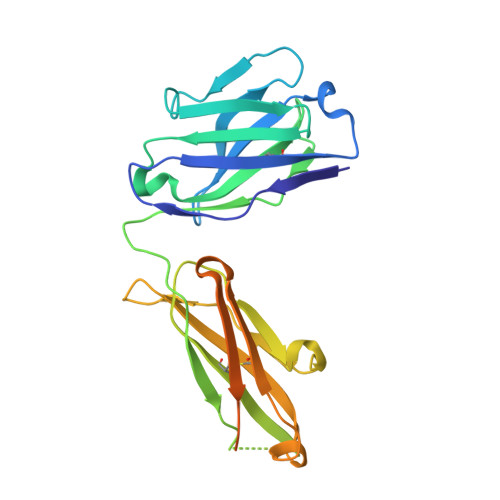
Crystal structures of human Fabs targeting the Bexsero meningococcal vaccine antigen NHBA.
Maritan, M., Cozzi, R., Lo Surdo, P., Veggi, D., Bottomley, M.J., Malito, E.(2017) Acta Crystallogr F Struct Biol Commun 73: 305-314
- PubMed: 28580917
- DOI: https://doi.org/10.1107/S2053230X17006021
- Primary Citation of Related Structures:
5N4G, 5N4J - PubMed Abstract:
Neisserial heparin-binding antigen (NHBA) is a surface-exposed lipoprotein from Neisseria meningitidis and is a component of the meningococcus B vaccine Bexsero. As part of a study to characterize the three-dimensional structure of NHBA and the molecular basis of the human immune response to Bexsero, the crystal structures of two fragment antigen-binding domains (Fabs) isolated from human monoclonal antibodies targeting NHBA were determined. Through a high-resolution analysis of the organization and the amino-acid composition of the CDRs, these structures provide broad insights into the NHBA epitopes recognized by the human immune system. As expected, these Fabs also show remarkable structural conservation, as shown by a structural comparison of 15 structures of apo Fab 10C3 which were obtained from crystals grown in different crystallization conditions and were solved while searching for a complex with a bound NHBA fragment or epitope peptide. This study also provides indirect evidence for the intrinsically disordered nature of two N-terminal regions of NHBA.
- GSK Vaccines, Via Fiorentina 1, 53100 Siena, Italy.
Organizational Affiliation: