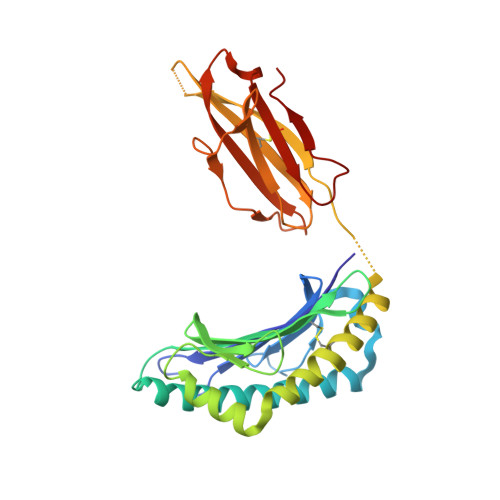
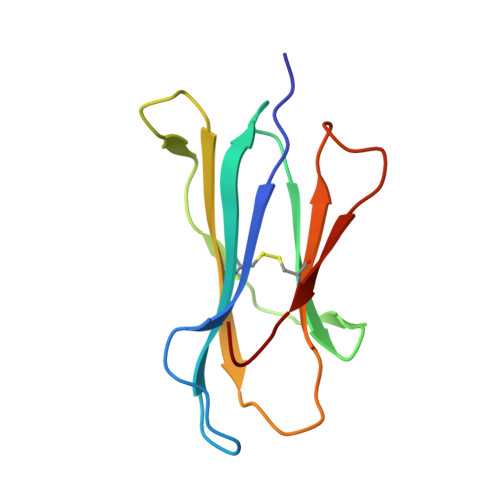
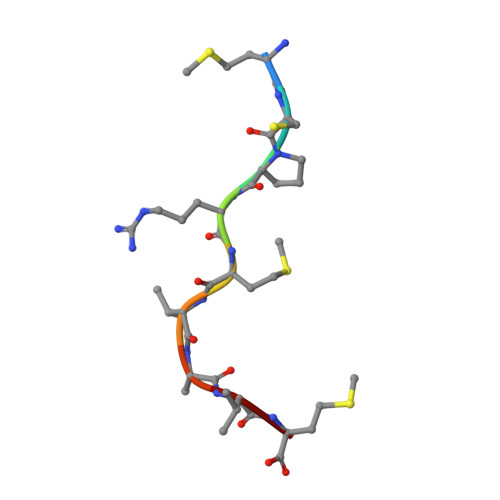
The Immunogenicity of a Proline-Substituted Altered Peptide Ligand toward the Cancer-Associated TEIPP Neoepitope Trh4 Is Unrelated to Complex Stability.
Hafstrand, I., Doorduijn, E.M., Sun, R., Talyzina, A., Sluijter, M., Pellegrino, S., Sandalova, T., Duru, A.D., van Hall, T., Achour, A.(2018) J Immunol 200: 2860-2868
- PubMed: 29507106
- DOI: https://doi.org/10.4049/jimmunol.1700228
- Primary Citation of Related Structures:
5MZM - PubMed Abstract:
Human cancers frequently display defects in Ag processing and presentation allowing for immune evasion, and they therefore constitute a significant challenge for T cell-based immunotherapy. We have previously demonstrated that the antigenicity of tumor-associated Ags can be significantly enhanced through unconventional residue modifications as a novel tool for MHC class I (MHC-I)-based immunotherapy approaches. We have also previously identified a novel category of cancer neo-epitopes, that is, T cell epitopes associated with impaired peptide processing (TEIPP), that are selectively presented by MHC-I on cells lacking the peptide transporter TAP. In this study, we demonstrate that substitution of the nonanchoring position 3 into a proline residue of the first identified TEIPP peptide, the murine Trh4, results in significantly enhanced recognition by antitumor CTLs toward the wild-type epitope. Although higher immunogenicity has in most cases been associated with increased MHC/peptide complex stability, our results demonstrate that the overall stability of H-2D b in complex with the highly immunogenic altered peptide ligand Trh4-p3P is significantly reduced compared with wild-type H-2D b /Trh4. Comparison of the crystal structures of the H-2D b /Trh4-p3P and H-2D b /Trh4 complexes revealed that the conformation of the nonconventional methionine anchor residue p5M is altered, deleting its capacity to form adequate sulfur-π interactions with H-2D b residues, thus reducing the overall longevity of the complex. Collectively, our results indicate that vaccination with Thr4-p3P significantly enhances T cell recognition of targets presenting the wild-type TEIPP epitope and that higher immunogenicity is not necessarily directly related to MHC/peptide complex stability, opening for the possibility to design novel peptide vaccines with reduced MHC/peptide complex stability.
- Science for Life Laboratory, Department of Medicine Solna, Karolinska Institutet, 17165 Stockholm, Sweden.
Organizational Affiliation: