Structural insights into chaperone addiction of toxin-antitoxin systems.
Guillet, V., Bordes, P., Bon, C., Marcoux, J., Gervais, V., Sala, A.J., Dos Reis, S., Slama, N., Mares-Mejia, I., Cirinesi, A.M., Maveyraud, L., Genevaux, P., Mourey, L.(2019) Nat Commun 10: 782-782
- PubMed: 30770830
- DOI: https://doi.org/10.1038/s41467-019-08747-4
- Primary Citation of Related Structures:
5MTW - PubMed Abstract:
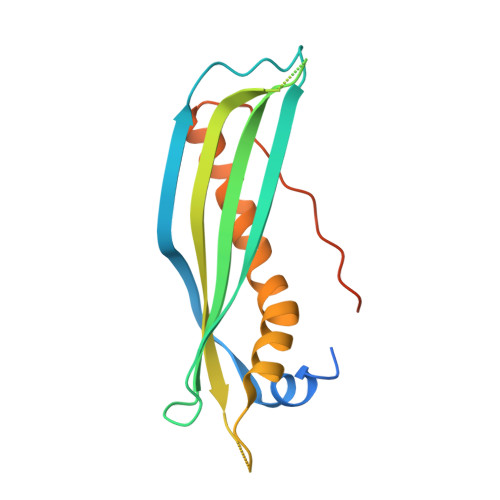
SecB chaperones assist protein export by binding both unfolded proteins and the SecA motor. Certain SecB homologs can also control toxin-antitoxin (TA) systems known to modulate bacterial growth in response to stress. In such TA-chaperone (TAC) systems, SecB assists the folding and prevents degradation of the antitoxin, thus facilitating toxin inhibition. Chaperone dependency is conferred by a C-terminal extension in the antitoxin known as chaperone addiction (ChAD) sequence, which makes the antitoxin aggregation-prone and prevents toxin inhibition. Using TAC of Mycobacterium tuberculosis, we present the structure of a SecB-like chaperone bound to its ChAD peptide. We find differences in the binding interfaces when compared to SecB-SecA or SecB-preprotein complexes, and show that the antitoxin can reach a functional form while bound to the chaperone. This work reveals how chaperones can use discrete surface binding regions to accommodate different clients or partners and thereby expand their substrate repertoire and functions.
- Institut de Pharmacologie et de Biologie Structurale, IPBS, Université de Toulouse, CNRS, UPS, 31077, Toulouse, France.
Organizational Affiliation: