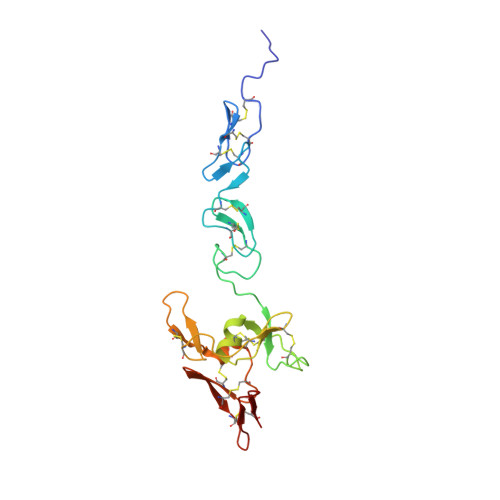
The N-Terminal Region of Fibrillin-1 Mediates a Bipartite Interaction with LTBP1.
Robertson, I.B., Dias, H.F., Osuch, I.H., Lowe, E.D., Jensen, S.A., Redfield, C., Handford, P.A.(2017) Structure 25: 1208-1221.e5
- PubMed: 28669633
- DOI: https://doi.org/10.1016/j.str.2017.06.003
- Primary Citation of Related Structures:
5MS9 - PubMed Abstract:
Fibrillin-1 (FBN1) mutations associated with Marfan syndrome lead to an increase in transforming growth factor β (TGF-β) activation in connective tissues resulting in pathogenic changes including aortic dilatation and dissection. Since FBN1 binds latent TGF-β binding proteins (LTBPs), the major reservoir of TGF-β in the extracellular matrix (ECM), we investigated the structural basis for the FBN1/LTBP1 interaction. We present the structure of a four-domain FBN1 fragment, EGF2-EGF3-Hyb1-cbEGF1 (FBN1 E2cbEGF1 ), which reveals a near-linear domain organization. Binding studies demonstrate a bipartite interaction between a C-terminal LTBP1 fragment and FBN1 E2cbEGF1 , which lies adjacent to the latency-associated propeptide (LAP)/TGF-β binding site of LTBP1. Modeling of the binding interface suggests that, rather than interacting along the longitudinal axis, LTBP1 anchors itself to FBN1 using two independent epitopes. As part of this mechanism, a flexible pivot adjacent to the FBN1/LTBP1 binding site allows LTBP1 to make contacts with different ECM networks while presumably facilitating a force-induced/traction-based TGF-β activation mechanism.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.
Organizational Affiliation: